| Glycosyl hydrolase family 14 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
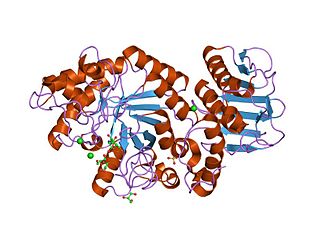
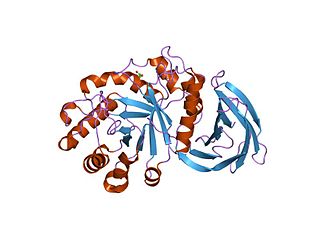
 beta-amylase from bacillus cereus var. mycoides in complex with maltose | |||||||||
| Identifiers | |||||||||
| Symbol | Glyco_hydro_14 | ||||||||
| Pfam | PF01373 | ||||||||
| Pfam clan | CL0058 | ||||||||
| InterPro | IPR001554 | ||||||||
| SCOP2 | 1byb / SCOPe / SUPFAM | ||||||||
| CAZy | GH14 | ||||||||
| |||||||||
In molecular biology, Glycoside hydrolase family 14 is a family of glycoside hydrolases.
Glycoside hydrolases EC 3.2.1. are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycoside hydrolases, based on sequence similarity, has led to the definition of >100 different families. [1] [2] [3] This classification is available on the CAZy web site, [4] [5] and also discussed at CAZypedia, an online encyclopedia of carbohydrate active enzymes. [6] [7]
Glycoside hydrolase family 14 CAZY GH_14 comprises enzymes with only one known activity; beta-amylase (EC 3.2.1.2). A Glu residue has been proposed as a catalytic residue, but it is not known if it is the nucleophile or the proton donor. Beta-amylase [8] [9] is an enzyme that hydrolyzes 1,4-alpha-glucosidic linkages in starch-type polysaccharide substrates so as to remove successive maltose units from the non-reducing ends of the chains. Beta-amylase is present in certain bacteria as well as in plants.
Three highly conserved sequence regions are found in all known beta-amylases. The first of these regions is located in the N-terminal section of the enzymes and contains an aspartate which is known [10] to be involved in the catalytic mechanism. The second, located in a more central location, is centred on a glutamate which is also involved [11] in the catalytic mechanism.
The 3D structure of a complex of soybean beta-amylase with an inhibitor (alpha-cyclodextrin) has been determined to 3.0 Å resolution by X-ray diffraction. [12] The enzyme folds into large and small domains: the large domain has a (beta alpha)8 super-secondary structural core, while the smaller is formed from two long loops extending from the beta-3 and beta-4 strands of the (beta alpha)8 fold. [12] The interface of the two domains, together with shorter loops from the (beta alpha)8 core, form a deep cleft, in which the inhibitor binds. [12] Two maltose molecules also bind in the cleft, one sharing a binding site with alpha-cyclodextrin, and the other sitting more deeply in the cleft. [12]