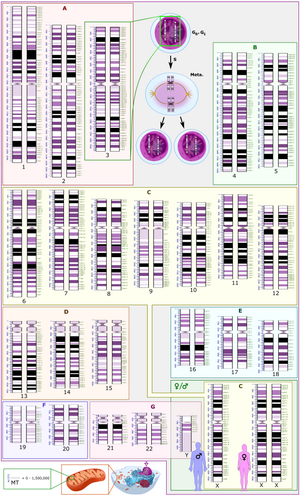
A karyotype is the general appearance of the complete set of chromosomes in the cells of a species or in an individual organism, mainly including their sizes, numbers, and shapes. Karyotyping is the process by which a karyotype is discerned by determining the chromosome complement of an individual, including the number of chromosomes and any abnormalities.

In genetics, a deletion is a mutation in which a part of a chromosome or a sequence of DNA is left out during DNA replication. Any number of nucleotides can be deleted, from a single base to an entire piece of chromosome. Some chromosomes have fragile spots where breaks occur which result in the deletion of a part of chromosome. The breaks can be induced by heat, viruses, radiations, chemicals. When a chromosome breaks, a part of it is deleted or lost, the missing piece of chromosome is referred to as deletion or a deficiency.

Cytogenetics is essentially a branch of genetics, but is also a part of cell biology/cytology, that is concerned with how the chromosomes relate to cell behaviour, particularly to their behaviour during mitosis and meiosis. Techniques used include karyotyping, analysis of G-banded chromosomes, other cytogenetic banding techniques, as well as molecular cytogenetics such as fluorescence in situ hybridization (FISH) and comparative genomic hybridization (CGH).

In genetics, chromosome translocation is a phenomenon that results in unusual rearrangement of chromosomes. This includes balanced and unbalanced translocation, with two main types: reciprocal-, and Robertsonian translocation. Reciprocal translocation is a chromosome abnormality caused by exchange of parts between non-homologous chromosomes. Two detached fragments of two different chromosomes are switched. Robertsonian translocation occurs when two non-homologous chromosomes get attached, meaning that given two healthy pairs of chromosomes, one of each pair "sticks" and blends together homogeneously.

Robertsonian translocation (ROB) is a chromosomal abnormality wherein a certain type of a chromosome becomes attached to another. It is the most common form of chromosomal translocation in humans, affecting 1 out of every 1,000 babies born. It does not usually cause health difficulties, but can in some cases result in genetic disorders such as Down syndrome and Patau syndrome. Robertsonian translocations result in a reduction in the number of chromosomes.

An inversion is a chromosome rearrangement in which a segment of a chromosome becomes inverted within its original position. An inversion occurs when a chromosome undergoes a two breaks within the chromosomal arm, and the segment between the two breaks inserts itself in the opposite direction in the same chromosome arm. The breakpoints of inversions often happen in regions of repetitive nucleotides, and the regions may be reused in other inversions. Chromosomal segments in inversions can be as small as 100 kilobases or as large as 100 megabases. The number of genes captured by an inversion can range from a handful of genes to hundreds of genes. Inversions can happen either through ectopic recombination, chromosomal breakage and repair, or non-homologous end joining.
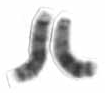
Chromosome 13 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 13 spans about 113 million base pairs and represents between 3.5 and 4% of the total DNA in cells.

Chromosome 22 is one of the 23 pairs of chromosomes in human cells. Humans normally have two copies of chromosome 22 in each cell. Chromosome 22 is the second smallest human chromosome, spanning about 51 million DNA base pairs and representing between 1.5 and 2% of the total DNA in cells.
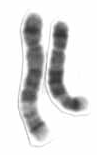
Chromosome 5 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 5 spans about 182 million base pairs and represents almost 6% of the total DNA in cells. Chromosome 5 is the 5th largest human chromosome, yet has one of the lowest gene densities. This is partially explained by numerous gene-poor regions that display a remarkable degree of non-coding and syntenic conservation with non-mammalian vertebrates, suggesting they are functionally constrained.
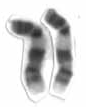
Chromosome 9 is one of the 23 pairs of chromosomes in humans. Humans normally have two copies of this chromosome, as they normally do with all chromosomes. Chromosome 9 spans about 150 million base pairs of nucleic acids and represents between 4.0 and 4.5% of the total DNA in cells.
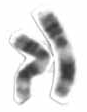
Chromosome 11 is one of the 23 pairs of chromosomes in humans. Humans normally have two copies of this chromosome. Chromosome 11 spans about 135 million base pairs and represents between 4 and 4.5 percent of the total DNA in cells. The shorter arm is termed 11p while the longer arm is 11q. At about 21.5 genes per megabase, chromosome 11 is one of the most gene-rich, and disease-rich, chromosomes in the human genome.

Chromosome 12 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 12 spans about 133 million base pairs and represents between 4 and 4.5 percent of the total DNA in cells.
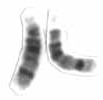
Chromosome 14 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 14 spans about 101 million base pairs and represents between 3 and 3.5% of the total DNA in cells.

Chromosome 15 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 15 spans about 99.7 million base pairs and represents between 3% and 3.5% of the total DNA in cells. Chromosome 15 is an acrocentric chromosome, with a very small short arm, which contains few protein coding genes among its 19 million base pairs. It has a larger long arm that is gene rich, spanning about 83 million base pairs.
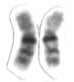
Chromosome 16 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 16 spans about 96 million base pairs and represents just under 3% of the total DNA in cells.

Chromosome 18 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 18 spans about 80 million base pairs and represents about 2.5 percent of the total DNA in cells.

Chromosome 20 is one of the 23 pairs of chromosomes in humans. Chromosome 20 spans around 66 million base pairs and represents between 2 and 2.5 percent of the total DNA in cells. Chromosome 20 was fully sequenced in 2001 and was reported to contain over 59 million base pairs. Since then, due to sequencing improvements and fixes, the length of chromosome 20 has been updated to just over 66 million base pairs.
A chromosomal abnormality, chromosomal anomaly, chromosomal aberration, chromosomal mutation, or chromosomal disorder, is a missing, extra, or irregular portion of chromosomal DNA. These can occur in the form of numerical abnormalities, where there is an atypical number of chromosomes, or as structural abnormalities, where one or more individual chromosomes are altered. Chromosome mutation was formerly used in a strict sense to mean a change in a chromosomal segment, involving more than one gene. Chromosome anomalies usually occur when there is an error in cell division following meiosis or mitosis. Chromosome abnormalities may be detected or confirmed by comparing an individual's karyotype, or full set of chromosomes, to a typical karyotype for the species via genetic testing.

A derivative chromosome (der) is a structurally rearranged chromosome generated either by a chromosome rearrangement involving two or more chromosomes or by multiple chromosome aberrations within a single chromosome. The term always refers to the chromosome that has an intact centromere. Derivative chromosomes are designated by the abbreviation der when used to describe a Karyotype. The derivative chromosome must be specified in parentheses followed by all aberrations involved in this derivative chromosome. The aberrations must be listed from pter to qter and not be separated by a comma.
The 2000s witnessed an explosion of genome sequencing and mapping in evolutionarily diverse species. While full genome sequencing of mammals is rapidly progressing, the ability to assemble and align orthologous whole chromosomal regions from more than a few species is not yet possible. The intense focus on the building of comparative maps for domestic, laboratory and agricultural (cattle) animals has traditionally been used to understand the underlying basis of disease-related and healthy phenotypes.