Related Research Articles

Orthohantavirus is a genus of single-stranded, enveloped, negative-sense RNA viruses in the family Hantaviridae within the order Bunyavirales. Members of this genus may be called orthohantaviruses or simply hantaviruses.

Bunyavirales is an order of segmented negative-strand RNA viruses with mainly tripartite genomes. Member viruses infect arthropods, plants, protozoans, and vertebrates. It is the only order in the class Ellioviricetes. The name Bunyavirales derives from Bunyamwera, where the original type species Bunyamwera orthobunyavirus was first discovered. Ellioviricetes is named in honor of late virologist Richard M. Elliott for his early work on bunyaviruses.

In infectious disease ecology and epidemiology, a natural reservoir, also known as a disease reservoir or a reservoir of infection, is the population of organisms or the specific environment in which an infectious pathogen naturally lives and reproduces, or upon which the pathogen primarily depends for its survival. A reservoir is usually a living host of a certain species, such as an animal or a plant, inside of which a pathogen survives, often without causing disease for the reservoir itself. By some definitions a reservoir may also be an environment external to an organism, such as a volume of contaminated air or water.
An emergent virus is a virus that is either newly appeared, notably increasing in incidence/geographic range or has the potential to increase in the near future. Emergent viruses are a leading cause of emerging infectious diseases and raise public health challenges globally, given their potential to cause outbreaks of disease which can lead to epidemics and pandemics. As well as causing disease, emergent viruses can also have severe economic implications. Recent examples include the SARS-related coronaviruses, which have caused the 2002–2004 outbreak of SARS (SARS-CoV-1) and the 2019–2023 pandemic of COVID-19 (SARS-CoV-2). Other examples include the human immunodeficiency virus, which causes HIV/AIDS; the viruses responsible for Ebola; the H5N1 influenza virus responsible for avian influenza; and H1N1/09, which caused the 2009 swine flu pandemic. Viral emergence in humans is often a consequence of zoonosis, which involves a cross-species jump of a viral disease into humans from other animals. As zoonotic viruses exist in animal reservoirs, they are much more difficult to eradicate and can therefore establish persistent infections in human populations.

Andes orthohantavirus (ANDV), a species of Orthohantavirus, is a major causative agent of hantavirus cardiopulmonary syndrome (HCPS) and hantavirus pulmonary syndrome (HPS) in South America. It is named for the Andes mountains of Chile and Argentina, where it was first discovered. Originating in the reservoir of rodents, Andes orthohantavirus is easily transmitted to humans who come into contact with infected rodents or their fecal droppings. However, infected rodents do not appear ill, so there is no readily apparent indicator to determine whether the rodent is infected or not. Additionally, Andes orthohantavirus, specifically, is the only hantavirus that can be spread by human to human contact via bodily fluids or long-term contact from one infected individual to a healthy person.
Black Creek Canal orthohantavirus (BCCV) is a single-stranded, negative sense RNA virus species of New World Orthohantavirus. It was first isolated in cotton rats found in the Black Creek Canal area of Dade County, Florida, in 1995. The discovery followed from an isolated case of Hantavirus pulmonary syndrome diagnosed in a Dade County resident.
Sangassou orthohantavirus(SANGV) is single-stranded, negative-sense RNA virus species of the genus Orthohantavirus in the Bunyavirales order. It was first isolated in an African wood mouse (Hylomyscus simus) in the forest in Guinea, West Africa in 2010. It is named for the village near where the mouse was trapped. It is the first indigenous Murinae-associated African hantavirus to be discovered.
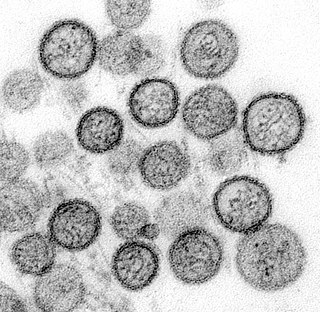
Hantavirus hemorrhagic fever with renal syndrome (HFRS) is a group of clinically similar illnesses caused by species of hantaviruses. It is also known as Korean hemorrhagic fever and epidemic hemorrhagic fever. It is found in Europe, Asia, and Africa. The species that cause HFRS include Hantaan orthohantavirus, Dobrava-Belgrade orthohantavirus, Saaremaa virus, Seoul orthohantavirus, Puumala orthohantavirus and other orthohantaviruses. Of these species, Hantaan River virus and Dobrava-Belgrade virus cause the most severe form of the syndrome and have the highest morbidity rates. When caused by the Puumala virus, it is also called nephropathia epidemica. This infection is known as sorkfeber in Swedish, myyräkuume in Finnish, and musepest in Norwegian.
Limestone Canyon virus (LSC) is a single-stranded, negative-sense RNA zoonotic Orthohantavirus that is genetically similar to Sin Nombre orthohantavirus which causes Hantavirus pulmonary syndrome (HPS) in humans. HPS causing hantaviruses are found only in the United States and South America.
Thottopalayam thottimvirus, formerly Thottapalayam virus, (TMPV) is single-stranded, enveloped, negative-sense RNA virus species of the genus Thottimvirus in the Bunyavirales order. It is the first hantavirus to be isolated from a shrew. It was discovered in India in 1964.
Topografov virus is an enveloped, negative-sense RNA virus of the genus Orthohantavirus in the Bunyavirales order. It is the first hantavirus to be isolated from Siberian lemmings found near the Topografov River in the Taymyr Peninsula, Siberia.
El Moro Canyon orthohantavirus is a single-stranded, negative sense RNA virus of the genus Orthohantavirus. It is a causative agent of Hantavirus pulmonary syndrome.
Nova virus is a single-stranded, negative-sense, enveloped RNA virus with a trisegmented genome. It belongs to one of the most divergent lineages of the hantavirus group, which consists of zoonotic viruses belonging to the family Bunyaviridae. As of now, no human cases of infection have been reported.
Rockport virus (RKPV) is a single-stranded, enveloped, negative-sense RNA orthohantavirus.
Asama orthohantavirus(ASAV), also called Asama virus, is a single-stranded, enveloped, segmented negative-sense RNA hantavirus. The hantavirus was isolated in Japan from Japanese shrew mole. Hantaviruses harbored by shrews are genetically closer to ASAV than to hantaviruses harbored by rodents. Host-switching may be evident in the future due to the viruses closeness to soricine shrew-borne hantaviruses. The detection of the ASAV was the first hantavirus found to be hosted by members of the family Talpidae, which includes shrew moles. Thoughts on hantavirus evolutionary history has expanded due to the discovery of ASAV.
Oxbow virus(OXBV) is a single-stranded, enveloped, negative-sense RNA orthohantavirus.
Thailand virus (THAIV) is a single-stranded, enveloped, negative-sense RNA orthohantavirus.
Gou virus (GOUV) is a single-stranded, negative-sense, enveloped novel RNA orthohantavirus. It is one of the known hantaviruses responsible for hantavirus hemorrhagic fever with renal syndrome in humans.

The bat virome is the group of viruses associated with bats. Bats host a diverse array of viruses, including all seven types described by the Baltimore classification system: (I) double-stranded DNA viruses; (II) single-stranded DNA viruses; (III) double-stranded RNA viruses; (IV) positive-sense single-stranded RNA viruses; (V) negative-sense single-stranded RNA viruses; (VI) positive-sense single-stranded RNA viruses that replicate through a DNA intermediate; and (VII) double-stranded DNA viruses that replicate through a single-stranded RNA intermediate. The greatest share of bat-associated viruses identified as of 2020 are of type IV, in the family Coronaviridae.
Blue River virus (BRV) is a single-stranded, negative sense RNA virus of New World hantavirus isolated from a white-footed mouse near the Blue River in Jackson County, Missouri in 1995. Its genome is similar to Sin Nombre orthohantavirus (SNV) but varies in the S1 and S2 segments. Like Sin Nombre orthohantavirus, Blue River virus causes Hantavirus pulmonary syndrome (HPS) in humans.
References
- ↑ Weiss S, Witkowski PT, Auste B, Nowak K, Weber N, Fahr J, et al. Hantavirus in bat, Sierra Leone [letter]. Emerg Infect Dis [serial on the Internet]. 2012 Jan
- ↑ Jung YT, Kim GR. Genomic characterization of M and S RNA segments of hantaviruses isolated from bats. Acta Virol. 1995;39:231–3.
- ↑ Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–45.
- ↑ Krüger DH, Schonrich G, Klempa B. Human pathogenic hantaviruses and prevention of infection. Hum Vaccin. 2011;7:685–93.
- ↑ Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–45. doi: 10.1128/CMR.00017-06. [PMC free article] [PubMed] [Cross Ref]
- ↑ Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O’Brien SJ Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–8. doi: 10.1038/35054550. [PubMed] [Cross Ref]