Protein targeting or protein sorting is the biological mechanism by which proteins are transported to their appropriate destinations within or outside the cell. Proteins can be targeted to the inner space of an organelle, different intracellular membranes, the plasma membrane, or to the exterior of the cell via secretion. Information contained in the protein itself directs this delivery process. Correct sorting is crucial for the cell; errors or dysfunction in sorting have been linked to multiple diseases.
Peptidoglycan or murein is a unique large macromolecule, a polysaccharide, consisting of sugars and amino acids that forms a mesh-like layer (sacculus) that surrounds the bacterial cytoplasmic membrane. The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is an oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer. Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer which is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply, a technique carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.

Ribosomes are macromolecular machines, found within all cells, that perform biological protein synthesis. Ribosomes link amino acids together in the order specified by the codons of messenger RNA molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA molecules and many ribosomal proteins. The ribosomes and associated molecules are also known as the translational apparatus.
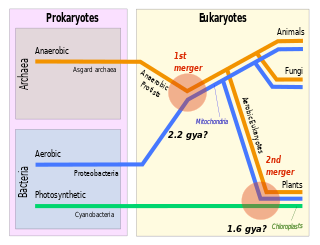
Symbiogenesis is the leading evolutionary theory of the origin of eukaryotic cells from prokaryotic organisms. The theory holds that mitochondria, plastids such as chloroplasts, and possibly other organelles of eukaryotic cells are descended from formerly free-living prokaryotes taken one inside the other in endosymbiosis. Mitochondria appear to be phylogenetically related to Rickettsiales bacteria, while chloroplasts are thought to be related to cyanobacteria.

Biochemistry is the study of the chemical processes in living organisms. It deals with the structure and function of cellular components such as proteins, carbohydrates, lipids, nucleic acids and other biomolecules.
The N-terminus (also known as the amino-terminus, NH2-terminus, N-terminal end or amine-terminus) is the start of a protein or polypeptide, referring to the free amine group (-NH2) located at the end of a polypeptide. Within a peptide, the amine group is bonded to the carboxylic group of another amino acid, making it a chain. That leaves a free carboxylic group at one end of the peptide, called the C-terminus, and a free amine group on the other end called the N-terminus. By convention, peptide sequences are written N-terminus to C-terminus, left to right (in LTR writing systems). This correlates the translation direction to the text direction, because when a protein is translated from messenger RNA, it is created from the N-terminus to the C-terminus, as amino acids are added to the carboxyl end of the protein.

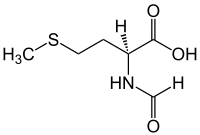
The start codon is the first codon of a messenger RNA (mRNA) transcript translated by a ribosome. The start codon always codes for methionine in eukaryotes and archaea and a N-formylmethionine (fMet) in bacteria, mitochondria and plastids.
Bacterial translation is the process by which messenger RNA is translated into proteins in bacteria.
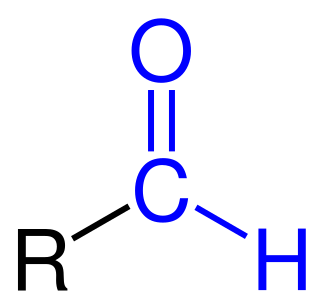
,Formylation refers to any chemical processes in which a compound is functionalized with a formyl group (-CH=O). In organic chemistry, the term is most commonly used with regards to aromatic compounds. In biochemistry the reaction is catalysed by enzymes such as formyltransferases.

Alexander J. Varshavsky is a Russian-American biochemist and geneticist. He works at the California Institute of Technology (Caltech) as the Morgan Professor of Biology. Varshavsky left Russia in 1977, emigrating to United States.
The formyl peptide receptors (FPR) belong to a class of G protein-coupled receptors involved in chemotaxis. In humans, there are three formyl peptide receptor isoforms, each encoded by a separate gene that are named FPR1, FPR2, and FPR3. These receptors were originally identified by their ability to bind N-formyl peptides such as N-formylmethionine produced by the degradation of either bacterial or host cells. Hence formyl peptide receptors are involved in mediating immune cell response to infection. These receptors may also act to suppress the immune system under certain conditions. The close phylogenetic relation of signaling in chemotaxis and olfaction was recently proved by detection formyl peptide receptor like proteins as a distinct family of vomeronasal organ chemosensors in mice.
Translocase is a general term for a protein that assists in moving another molecule, usually across a cell membrane. These enzymes catalyze the movement of ions or molecules across membranes or their separation within membranes. The reaction is designated as a transfer from “side 1” to “side 2” because the designations “in” and “out”, which had previously been used, can be ambiguous. Translocases are the most common secretion system in Gram positive bacteria.

N-formyl peptide receptor 2 (FPR2) is a G-protein coupled receptor (GPCR) located on the surface of many cell types of various animal species. The human receptor protein is encoded by the FPR2 gene and is activated to regulate cell function by binding any one of a wide variety of ligands including not only certain N-Formylmethionine-containing oligopeptides such as N-Formylmethionine-leucyl-phenylalanine (FMLP) but also the polyunsaturated fatty acid metabolite of arachidonic acid, lipoxin A4 (LXA4). Because of its interaction with lipoxin A4, FPR2 is also commonly named the ALX/FPR2 or just ALX receptor.

N-Formylmethionyl-leucyl-phenylalanine is an N-formylated tripeptide and sometimes simply referred to as chemotactic peptide is a potent polymorphonuclear leukocyte (PMN) chemotactic factor and is also a macrophage activator.

Methionine aminopeptidase 2 is an enzyme that in humans is encoded by the METAP2 gene.

N-formyl peptide receptor 3 (FPR3) is a receptor protein that in humans is encoded by the FPR3 gene.
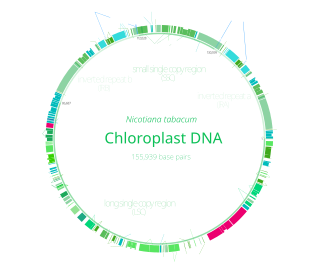
Chloroplast DNA (cpDNA), also known as plastid DNA (ptDNA) is the DNA located in chloroplasts, which are photosynthetic organelles located within the cells of some eukaryotic organisms. Chloroplasts, like other types of plastid, contain a genome separate from that in the cell nucleus. The existence of chloroplast DNA was identified biochemically in 1959, and confirmed by electron microscopy in 1962. The discoveries that the chloroplast contains ribosomes and performs protein synthesis revealed that the chloroplast is genetically semi-autonomous. The first complete chloroplast genome sequences were published in 1986, Nicotiana tabacum (tobacco) by Sugiura and colleagues and Marchantia polymorpha (liverwort) by Ozeki et al. Since then, tens of thousands of chloroplast genomes from various species have been sequenced.
The N-end rule is a rule that governs the rate of protein degradation through recognition of the N-terminal residue of proteins. The rule states that the N-terminal amino acid of a protein determines its half-life. The rule applies to both eukaryotic and prokaryotic organisms, but with different strength, rules, and outcome. In eukaryotic cells, these N-terminal residues are recognized and targeted by ubiquitin ligases, mediating ubiquitination thereby marking the protein for degradation. The rule was initially discovered by Alexander Varshavsky and co-workers in 1986. However, only rough estimations of protein half-life can be deduced from this 'rule', as N-terminal amino acid modification can lead to variability and anomalies, whilst amino acid impact can also change from organism to organism. Other degradation signals, known as degrons, can also be found in sequence.

Formyl peptide receptor 1 is a cell surface receptor protein that in humans is encoded by the formyl peptide receptor 1 (FPR1) gene. This gene encodes a G protein-coupled receptor cell surface protein that binds and is activated by N-Formylmethionine-containing oligopeptides, particularly N-Formylmethionine-leucyl-phenylalanine (FMLP). FPR1 is prominently expressed by mammalian phagocytic and blood leukocyte cells where it functions to mediate these cells' responses to the N-formylmethionine-containing oligopeptides which are released by invading microorganisms and injured tissues. FPR1 directs these cells to sites of invading pathogens or disrupted tissues and then stimulates these cells to kill the pathogens or to remove tissue debris; as such, it is an important component of the innate immune system that operates in host defense and damage control.
Thimet oligopeptidases, also known as TOPs, are a type of M3 metallopeptidases. These enzymes can be found in animals and plants, showing distinctive functions. In animals and humans, they are involved in the degradation of peptides, such as bradykinin, neurotensin, angiotensin I, and Aβ peptide, helping to regulate physiological processes. In plants, their role is related to the degradation of targeting peptides and the immune response to pathogens through Salicylic Acid (SA)-dependent stress signaling. In Arabidopsis thaliana—recognized as a model plant for scientific studies—two thimet oligopeptidases, known as TOP1 and TOP2, have been identified as targets for salicylic acid binding in the plant. These TOP enzymes are key components to understand the SA-mediated signaling where interactions exist with different components and most of the pathways are unknown.