Hydrogen peroxide is a chemical compound with the formula H2O2. In its pure form, it is a very pale blue liquid that is slightly more viscous than water. It is used as an oxidizer, bleaching agent, and antiseptic, usually as a dilute solution in water for consumer use and in higher concentrations for industrial use. Concentrated hydrogen peroxide, or "high-test peroxide", decomposes explosively when heated and has been used as both a monopropellant and an oxidizer in rocketry.
Catalase is a common enzyme found in nearly all living organisms exposed to oxygen which catalyzes the decomposition of hydrogen peroxide to water and oxygen. It is a very important enzyme in protecting the cell from oxidative damage by reactive oxygen species (ROS). Catalase has one of the highest turnover numbers of all enzymes; one catalase molecule can convert millions of hydrogen peroxide molecules to water and oxygen each second.
A dehydrogenase is an enzyme belonging to the group of oxidoreductases that oxidizes a substrate by reducing an electron acceptor, usually NAD+/NADP+ or a flavin coenzyme such as FAD or FMN. Like all catalysts, they catalyze reverse as well as forward reactions, and in some cases this has physiological significance: for example, alcohol dehydrogenase catalyzes the oxidation of ethanol to acetaldehyde in animals, but in yeast it catalyzes the production of ethanol from acetaldehyde.

The glucose oxidase enzyme also known as notatin is an oxidoreductase that catalyses the oxidation of glucose to hydrogen peroxide and D-glucono-δ-lactone. This enzyme is produced by certain species of fungi and insects and displays antibacterial activity when oxygen and glucose are present.
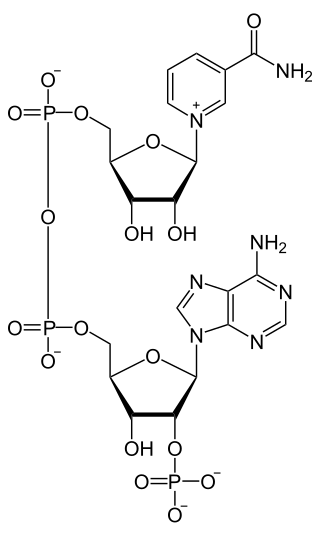
Nicotinamide adenine dinucleotide phosphate, abbreviated NADP+ or, in older notation, TPN (triphosphopyridine nucleotide), is a cofactor used in anabolic reactions, such as the Calvin cycle and lipid and nucleic acid syntheses, which require NADPH as a reducing agent ('hydrogen source'). NADPH is the reduced form, whereas NADP+ is the oxidized form. NADP+ is used by all forms of cellular life.
Respiratory burst is the rapid release of the reactive oxygen species (ROS), superoxide anion and hydrogen peroxide, from different cell types.
Any enzyme system that includes cytochrome P450 protein or domain can be called a P450-containing system.
Ascorbate peroxidase (or L-ascorbate peroxidase, APX or APEX) (EC 1.11.1.11) is an enzyme that catalyzes the chemical reaction

4-hydroxyphenylacetate 3-monooxygenase (EC 1.14.14.9) is an enzyme that catalyzes the chemical reaction
In enzymology, a malate oxidase (EC 1.1.3.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a fatty-acid peroxidase (EC 1.11.1.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a lignin peroxidase (EC 1.11.1.14) is an enzyme that catalyzes the chemical reaction
In enzymology, a manganese peroxidase (EC 1.11.1.13) is an enzyme that catalyzes the chemical reaction
In enzymology, a NADPH peroxidase (EC 1.11.1.2) is an enzyme that catalyzes the chemical reaction

Renalase, FAD-dependent amine oxidase is an enzyme that in humans is encoded by the RNLS gene. Renalase is a flavin adenine dinucleotide-dependent amine oxidase that is secreted into the blood from the kidney.
Hypothiocyanite is the anion [OSCN]− and the conjugate base of hypothiocyanous acid (HOSCN). It is an organic compound part of the thiocyanates as it contains the functional group SCN. It is formed when an oxygen is singly bonded to the thiocyanate group. Hypothiocyanous acid is a fairly weak acid; its acid dissociation constant (pKa) is 5.3.

Lactoperoxidase is a peroxidase enzyme secreted from mammary, salivary and other mucosal glands including the lungs, bronchii and nose that functions as a natural and the first line of defense against bacteria and viruses. Lactoperoxidase is a member of the heme peroxidase family of enzymes. In humans, lactoperoxidase is encoded by the LPO gene.
Haem peroxidases (or heme peroxidases) are haem-containing enzymes that use hydrogen peroxide as the electron acceptor to catalyse a number of oxidative reactions. Most haem peroxidases follow the reaction scheme:

A selenenic acid is an organoselenium compound and an oxoacid with the general formula RSeOH, where R ≠ H. It is the first member of the family of organoselenium oxoacids, which also include seleninic acids and selenonic acids, which are RSeO2H and RSeO3H, respectively. Selenenic acids derived from selenoenzymes are thought to be responsible for the antioxidant activity of these enzymes. This functional group is sometimes called SeO-selenoperoxol.

NADH:ubiquinone reductase (non-electrogenic) (EC 1.6.5.9, NDH-2, ubiquinone reductase, coenzyme Q reductase, dihydronicotinamide adenine dinucleotide-coenzyme Q reductase, DPNH-coenzyme Q reductase, DPNH-ubiquinone reductase, NADH-coenzyme Q oxidoreductase, NADH-coenzyme Q reductase, NADH-CoQ oxidoreductase, NADH-CoQ reductase) is an enzyme with systematic name NADH:ubiquinone oxidoreductase. This enzyme catalyses the following chemical reaction: