
The cytochrome complex, or cyt c, is a small hemeprotein found loosely associated with the inner membrane of the mitochondrion where it plays a critical role in cellular respiration. It transfers electrons between Complexes III and IV. Cytochrome c is highly water-soluble, unlike other cytochromes. It is capable of undergoing oxidation and reduction as its iron atom converts between the ferrous and ferric forms, but does not bind oxygen. It also plays a major role in cell apoptosis. In humans, cytochrome c is encoded by the CYCS gene.

Leghemoglobin is an oxygen-carrying phytoglobin found in the nitrogen-fixing root nodules of leguminous plants. It is produced by these plants in response to the roots being colonized by nitrogen-fixing bacteria, termed rhizobia, as part of the symbiotic interaction between plant and bacterium: roots not colonized by Rhizobium do not synthesise leghemoglobin. Leghemoglobin has close chemical and structural similarities to hemoglobin, and, like hemoglobin, is red in colour. It was originally thought that the heme prosthetic group for plant leghemoglobin was provided by the bacterial symbiont within symbiotic root nodules. However, subsequent work shows that the plant host strongly expresses heme biosynthesis genes within nodules, and that activation of those genes correlates with leghemoglobin gene expression in developing nodules.

Heme, or haem, is a precursor to hemoglobin, which is necessary to bind oxygen in the bloodstream. Heme is biosynthesized in both the bone marrow and the liver.

A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst. Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound.

Cytochromes P450 are a superfamily of enzymes containing heme as a cofactor that mostly, but not exclusively, function as monooxygenases. In mammals, these proteins oxidize steroids, fatty acids, and xenobiotics, and are important for the clearance of various compounds, as well as for hormone synthesis and breakdown. In 1963, Estabrook, Cooper, and Rosenthal described the role of CYP as a catalyst in steroid hormone synthesis and drug metabolism. In plants, these proteins are important for the biosynthesis of defensive compounds, fatty acids, and hormones.

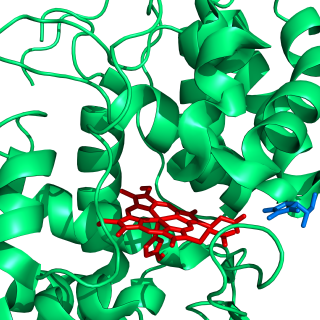
Lipoxygenases (LOX) are a family of (non-heme) iron-containing enzymes most of which catalyze the dioxygenation of polyunsaturated fatty acids in lipids containing a cis,cis-1,4-pentadiene into cell signaling agents that serve diverse roles as autocrine signals that regulate the function of their parent cells, paracrine signals that regulate the function of nearby cells, and endocrine signals that regulate the function of distant cells.

Heme oxygenase, or haem oxygenase, is an enzyme that catalyzes the degradation of heme to produce biliverdin, ferrous ion, and carbon monoxide.

Biliverdin reductase (BVR) is an enzyme found in all tissues under normal conditions, but especially in reticulo-macrophages of the liver and spleen. BVR facilitates the conversion of biliverdin to bilirubin via the reduction of a double-bond between the second and third pyrrole ring into a single-bond.
Ascorbate peroxidase (or L-ascorbate peroxidase, APX or APEX) (EC 1.11.1.11) is an enzyme that catalyzes the chemical reaction

Malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) (EC 1.1.1.40) or NADP-malic enzyme (NADP-ME) is an enzyme that catalyzes the chemical reaction in the presence of a bivalent metal ion:

The enzyme 4-hydroxybenzoate 3-monooxygenase, also commonly referred to as para-hydroxybenzoate hydroxylase (PHBH), is a flavoprotein belonging to the family of oxidoreductases. Specifically, it is a hydroxylase, and is one of the most studied enzymes and catalyzes reactions involved in soil detoxification, metabolism, and other biosynthetic processes.

α-Parinaric acid is a conjugated polyunsaturated fatty acid. Discovered by Tsujimoto and Koyanagi in 1933, it contains 18 carbon atoms and 4 conjugated double bonds. The repeating single bond-double bond structure of α-parinaric acid distinguishes it structurally and chemically from the usual "methylene-interrupted" arrangement of polyunsaturated fatty acids that have double-bonds and single bonds separated by a methylene unit (−CH2−). Because of the fluorescent properties conferred by the alternating double bonds, α-parinaric acid is commonly used as a molecular probe in the study of biomembranes.
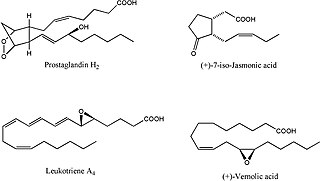
Oxylipins constitute a family of oxygenated natural products which are formed from fatty acids by pathways involving at least one step of dioxygen-dependent oxidation. Oxylipins are derived from polyunsaturated fatty acids (PUFAs) by COX enzymes (cyclooxygenases), by LOX enzymes (lipoxygenases), or by cytochrome P450 epoxygenase.
Fatty-acid peroxygenase is an enzyme with systematic name fatty acid:hydroperoxide oxidoreductase (RH-hydroxylating). This enzyme catalyses the following chemical reaction
Linoleate 8R-lipoxygenase (EC 1.13.11.60, linoleic acid 8R-dioxygenase, 5,8-LDS (bifunctional enzyme), 7,8-LDS (bifunctional enzyme), 5,8-linoleate diol synthase (bifunctional enzyme), 7,8-linoleate diol synthase (bifunctional enzyme), PpoA) is an enzyme with systematic name linoleate:oxygen (8R)-oxidoreductase. This enzyme catalyses the following chemical reaction
Colneleate synthase (EC 4.2.1.121, 9-divinyl ether synthase, 9-DES, CYP74D, CYP74D1, CYP74 cytochrome P-450, DES1) is an enzyme with systematic name (8E)-9-((1E,3E)-nona-1,3-dien-1-yloxy)non-8-enoate synthase. This enzyme catalyses the following chemical reaction
9,12-octadecadienoate 8-hydroperoxide 8R-isomerase is an enzyme with systematic name (8R,9Z,12Z)-8-hydroperoxyoctadeca-9,12-dienoate hydroxymutase ( -5,8-dihydroxyoctadeca-9,12-dienoate-forming). This enzyme catalyses the following chemical reaction
9,12-octadecadienoate 8-hydroperoxide 8S-isomerase is an enzyme with systematic name (8R,9Z,12Z)-8-hydroperoxyoctadeca-9,12-dienoate hydroxymutase ( -7,8-dihydroxyoctadeca-9,12-dienoate-forming). This enzyme catalyses the following chemical reaction
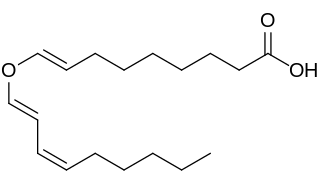
Divinylether fatty acids contain a fatty acid chemically combined with a doubly unsaturated carbon chain linked by an oxygen atom (ether). Fatty acid hydroperoxides generated by plant lipoxygenases from linoleic and linolenic acids are known to serve as substrates for a divinyl ether synthase which produces divinyl ether fatty acids. Up to date divinyl ethers were detected only within the plant kingdom.

Hydroperoxide lyases are enzymes that catalyze the cleavage of C-C bonds in the hydroperoxides of fatty acids. They belong to the cytochrome P450 enzyme family.















