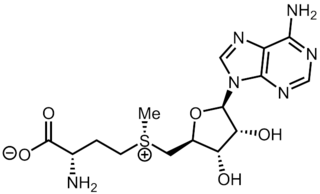
S-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952.

Base excision repair (BER) is a cellular mechanism, studied in the fields of biochemistry and genetics, that repairs damaged DNA throughout the cell cycle. It is responsible primarily for removing small, non-helix-distorting base lesions from the genome. The related nucleotide excision repair pathway repairs bulky helix-distorting lesions. BER is important for removing damaged bases that could otherwise cause mutations by mispairing or lead to breaks in DNA during replication. BER is initiated by DNA glycosylases, which recognize and remove specific damaged or inappropriate bases, forming AP sites. These are then cleaved by an AP endonuclease. The resulting single-strand break can then be processed by either short-patch or long-patch BER.
Photolyases are DNA repair enzymes that repair damage caused by exposure to ultraviolet light. These enzymes require visible light both for their own activation and for the actual DNA repair. The DNA repair mechanism involving photolyases is called photoreactivation. They mainly convert pyrimidine dimers into a normal pair of pyrimidine bases.
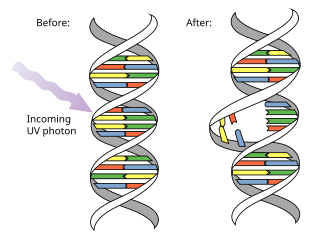
Pyrimidine dimers are molecular lesions formed from thymine or cytosine bases in DNA via photochemical reactions, commonly associated with direct DNA damage. Ultraviolet light induces the formation of covalent linkages between consecutive bases along the nucleotide chain in the vicinity of their carbon–carbon double bonds. The photo-coupled dimers are fluorescent. The dimerization reaction can also occur among pyrimidine bases in dsRNA —uracil or cytosine. Two common UV products are cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts. These premutagenic lesions alter the structure of the DNA helix and cause non-canonical base pairing. Specifically, adjacent thymines or cytosines in DNA will form a cyclobutane ring when joined together and cause a distortion in the DNA. This distortion prevents replication or transcription machinery beyond the site of the dimerization. Up to 50–100 such reactions per second might occur in a skin cell during exposure to sunlight, but are usually corrected within seconds by photolyase reactivation or nucleotide excision repair. In humans, the most common form of DNA repair is nucleotide excision repair (NER). In contrast, organisms such as bacteria can counterintuitively harvest energy from the sun to fix DNA damage from pyrimidine dimers via photolyase activity. If these lesions are not fixed, polymerase machinery may misread or add in the incorrect nucleotide to the strand. If the damage to the DNA is overwhelming, mutations can arise within the genome of an organism and may lead to the production of cancer cells. Uncorrected lesions can inhibit polymerases, cause misreading during transcription or replication, or lead to arrest of replication. It causes sunburn and it triggers the production of melanin. Pyrimidine dimers are the primary cause of melanomas in humans.

DNA damage-binding protein 2 is a protein that in humans is encoded by the DDB2 gene.

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction

The enzyme methionine γ-lyase (EC 4.4.1.11, MGL) is in the γ-family of PLP-dependent enzymes. It degrades sulfur-containing amino acids to α-keto acids, ammonia, and thiols:
Biotin synthase (BioB) is an enzyme that catalyzes the conversion of dethiobiotin (DTB) to biotin; this is the final step in the biotin biosynthetic pathway. Biotin, also known as vitamin B7, is a cofactor used in carboxylation, decarboxylation, and transcarboxylation reactions in many organisms including humans. Biotin synthase is an S-Adenosylmethionine (SAM) dependent enzyme that employs a radical mechanism to thiolate dethiobiotin, thus converting it to biotin.
In enzymology, a pinene synthase is an enzyme that catalyzes the chemical reaction

DNA-3-methyladenine glycosylase also known as 3-alkyladenine DNA glycosylase (AAG) or N-methylpurine DNA glycosylase (MPG) is an enzyme that in humans is encoded by the MPG gene.

Endonuclease III-like protein 1 is an enzyme that in humans is encoded by the NTHL1 gene.
Endonuclease VIII-like 2 is an enzyme that in humans is encoded by the NEIL2 gene.
Spore photoproduct lyase is a radical SAM enzyme that repairs DNA cross linking of thymine bases caused by UV-radiation. There are several types of thymine cross linking, but SPL specifically targets 5-thyminyl-5,6-dihydrothymine, which is also called spore photoproduct (SP). Spore photoproduct is the predominant type of thymine crosslinking in germinating endospores, which is why SPL is unique to organisms that produce endospores, such as Bacillus subtilis. Other types of thymine crosslinking, such as cyclobutane pyrimidine dimers (CPD) and pyrimidine (6-4) pyrimidone photoproducts (6-4PPs), are less commonly formed in endospores. These differences in DNA crosslinking are a function of differing DNA structure. Spore genomic DNA features many DNA binding proteins called small acid soluble proteins, which changes the DNA from the traditional B-form conformation to an A-form conformation. This difference in conformation is believed to be the reason why dormant spores predominantly accumulate SP in response to UV-radiation, rather than other forms of cross linking. Spores cannot repair cross-linking while dormant, instead the SPs are repaired during germination to allow the vegetative cell to function normally. When not repaired, spore photoproduct and other types of crosslinking can cause mutations by blocking transcription and replication past the point of the crosslinking. The repair mechanism utilizing spore photoproduct lyase is one of the reasons for the resilience of certain bacterial spores.

In molecular biology, the H2TH domain is a DNA-binding domain found in DNA glycosylase/AP lyase enzymes, which are involved in base excision repair of DNA damaged by oxidation or by mutagenic agents. Most damage to bases in DNA is repaired by the base excision repair pathway. These enzymes are primarily from bacteria, and have both DNA glycosylase activity EC 3.2.2.- and AP lyase activity EC 4.2.99.18. Examples include formamidopyrimidine-DNA glycosylases and endonuclease VIII (Nei).
Radical SAM is a designation for a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. rSAMs comprise the largest superfamily of metal-containing enzymes.
[Fructose-bisphosphate aldolase]-lysine N-methyltransferase (EC 2.1.1.259) is an enzyme that catalyses the following chemical reaction
Quinolinate synthase (EC 2.5.1.72, NadA, QS, quinolinate synthetase) is an enzyme with systematic name glycerone phosphate:iminosuccinate alkyltransferase (cyclizing). This enzyme catalyses the following chemical reaction
(6-4)DNA photolyase is an enzyme with systematic name (6-4) photoproduct pyrimidine-lyase. This enzyme catalyses the following chemical reaction
Methylthiotransferases are enzymes of the radical S-adenosyl methionine superfamily. These enzymes catalyze the addition of a methylthio group to various biochemical compounds including tRNA and proteins. Methylthiotransferases are classified into one of four classes based on their substrates and mechanisms. All methylthiotransferases have been shown to contain two Fe-S clusters, one canonical cluster and one auxiliary cluster, that both function in the addition of the methylthio group to the substrate.

5,6-Dihydro-5(α-thyminyl)thymine is a DNA pyrimidine dimer photoproduct produced when DNA in bacterial spores is exposed to ultraviolet light. In bacteria, this DNA base dimer deforms the structure of DNA, so endospore forming bacteria have an enzyme called spore photoproduct lyase that repairs this damage.













