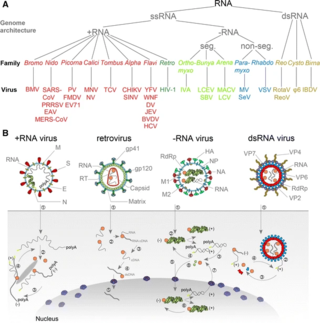
An RNA virus is a virus—other than a retrovirus—that has ribonucleic acid (RNA) as its genetic material. The nucleic acid is usually single-stranded RNA (ssRNA) but it may be double-stranded (dsRNA). Notable human diseases caused by RNA viruses include the common cold, influenza, SARS, MERS, COVID-19, Dengue Virus, hepatitis C, hepatitis E, West Nile fever, Ebola virus disease, rabies, polio, mumps, and measles.

Picornaviruses are a group of related nonenveloped RNA viruses which infect vertebrates including fish, mammals, and birds. They are viruses that represent a large family of small, positive-sense, single-stranded RNA viruses with a 30 nm icosahedral capsid. The viruses in this family can cause a range of diseases including the common cold, poliomyelitis, meningitis, hepatitis, and paralysis.

The mumps virus (MuV) is the virus that causes mumps. MuV contains a single-stranded, negative-sense genome made of ribonucleic acid (RNA). Its genome is about 15,000 nucleotides in length and contains seven genes that encode nine proteins. The genome is encased by a capsid that is in turn surrounded by a viral envelope. MuV particles, called virions, are pleomorphic in shape and vary in size from 100 to 600 nanometers in diameter. One serotype and twelve genotypes that vary in their geographic distribution are recognized. Humans are the only natural host of the mumps virus.

DNA sequencing is the process of determining the nucleic acid sequence – the order of nucleotides in DNA. It includes any method or technology that is used to determine the order of the four bases: adenine, guanine, cytosine, and thymine. The advent of rapid DNA sequencing methods has greatly accelerated biological and medical research and discovery.

Metagenomics is the study of genetic material recovered directly from environmental or clinical samples by a method called sequencing. The broad field may also be referred to as environmental genomics, ecogenomics, community genomics or microbiomics.
Baltimore classification is a system used to classify viruses based on their manner of messenger RNA (mRNA) synthesis. By organizing viruses based on their manner of mRNA production, it is possible to study viruses that behave similarly as a distinct group. Seven Baltimore groups are described that take into consideration whether the viral genome is made of deoxyribonucleic acid (DNA) or ribonucleic acid (RNA), whether the genome is single- or double-stranded, and whether the sense of a single-stranded RNA genome is positive or negative.

RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.
Marnaviridae is a family of positive-stranded RNA viruses in the order Picornavirales that infect various photosynthetic marine protists. Members of the family have non-enveloped, icosahedral capsids. Replication occurs in the cytoplasm and causes lysis of the host cell. The first species of this family that was isolated is Heterosigma akashiwo RNA virus (HaRNAV) in the genus Marnavirus, which infects the toxic bloom-forming Raphidophyte alga, Heterosigma akashiwo. As of 2021, there are twenty species across seven genera in this family, as well as many other related virus sequences discovered through metagenomic sequencing that are currently unclassified.
Picobirnavirus is a genus of double-stranded RNA viruses. It is the only genus in the family Picobirnaviridae. Although amniotes, especially mammals, were thought to serve as hosts, it has been recently suggested that these viruses might infect bacteria and possibly some other invertebrates. If they do infect bacteria, then they are Bacteriophages. There are three species in this genus. Associated symptoms include gastroenteritis in animals and humans, though the disease association is unclear.

Reverse genetics is a method in molecular genetics that is used to help understand the function(s) of a gene by analysing the phenotypic effects caused by genetically engineering specific nucleic acid sequences within the gene. The process proceeds in the opposite direction to forward genetic screens of classical genetics. While forward genetics seeks to find the genetic basis of a phenotype or trait, reverse genetics seeks to find what phenotypes are controlled by particular genetic sequences.

Viral metagenomics uses metagenomic technologies to detect viral genomic material from diverse environmental and clinical samples. Viruses are the most abundant biological entity and are extremely diverse; however, only a small fraction of viruses have been sequenced and only an even smaller fraction have been isolated and cultured. Sequencing viruses can be challenging because viruses lack a universally conserved marker gene so gene-based approaches are limited. Metagenomics can be used to study and analyze unculturable viruses and has been an important tool in understanding viral diversity and abundance and in the discovery of novel viruses. For example, metagenomics methods have been used to describe viruses associated with cancerous tumors and in terrestrial ecosystems.
Narnavirus is a genus of positive-strand RNA viruses in the family Narnaviridae. Fungi serve as natural hosts. There are two species in this genus. Member viruses have been shown to be required for sexual reproduction of Rhizopus microsporus. Narnaviruses have a naked RNA genome without a virion and derive their name from this feature.

Virome refers to the assemblage of viruses that is often investigated and described by metagenomic sequencing of viral nucleic acids that are found associated with a particular ecosystem, organism or holobiont. The word is frequently used to describe environmental viral shotgun metagenomes. Viruses, including bacteriophages, are found in all environments, and studies of the virome have provided insights into nutrient cycling, development of immunity, and a major source of genes through lysogenic conversion. Also, the human virome has been characterized in nine organs of 31 Finnish individuals using qPCR and NGS methodologies.

Positive-strand RNA viruses are a group of related viruses that have positive-sense, single-stranded genomes made of ribonucleic acid. The positive-sense genome can act as messenger RNA (mRNA) and can be directly translated into viral proteins by the host cell's ribosomes. Positive-strand RNA viruses encode an RNA-dependent RNA polymerase (RdRp) which is used during replication of the genome to synthesize a negative-sense antigenome that is then used as a template to create a new positive-sense viral genome.

The first step of transcription for some negative, single-stranded RNA viruses is cap snatching, in which the first 10 to 20 residues of a host cell RNA are removed (snatched) and used as the 5′ cap and primer to initiate the synthesis of the nascent viral mRNA. The viral RNA-dependent RNA polymerase (RdRp) can then proceed to replicate the negative-sense genome from the positive-sense template. Cap-snatching also explains why some viral mRNA have 5’ terminal extensions of 10-20 nucleotides that are not encoded for in the genome. Examples of viruses that engage in cap-snatching include influenza viruses (Orthomyxoviridae), Lassa virus (Arenaviridae), hantaan virus (Hantaviridae) and rift valley fever virus (Phenuiviridae). Most viruses snatch 15-20 nucleotides except for the families Arenaviridae and Nairoviridae and the genus Thogotovirus (Orthomyxoviridae) which use a shorter strand.
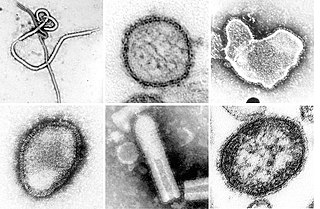
Negative-strand RNA viruses are a group of related viruses that have negative-sense, single-stranded genomes made of ribonucleic acid (RNA). They have genomes that act as complementary strands from which messenger RNA (mRNA) is synthesized by the viral enzyme RNA-dependent RNA polymerase (RdRp). During replication of the viral genome, RdRp synthesizes a positive-sense antigenome that it uses as a template to create genomic negative-sense RNA. Negative-strand RNA viruses also share a number of other characteristics: most contain a viral envelope that surrounds the capsid, which encases the viral genome, −ssRNA virus genomes are usually linear, and it is common for their genome to be segmented.

Riboviria is a realm of viruses that includes all viruses that use a homologous RNA-dependent polymerase for replication. It includes RNA viruses that encode an RNA-dependent RNA polymerase, as well as reverse-transcribing viruses that encode an RNA-dependent DNA polymerase. RNA-dependent RNA polymerase (RdRp), also called RNA replicase, produces RNA from RNA. RNA-dependent DNA polymerase (RdDp), also called reverse transcriptase (RT), produces DNA from RNA. These enzymes are essential for replicating the viral genome and transcribing viral genes into messenger RNA (mRNA) for translation of viral proteins.

Orthornavirae is a kingdom of viruses that have genomes made of ribonucleic acid (RNA), including genes which encode an RNA-dependent RNA polymerase (RdRp). The RdRp is used to transcribe the viral RNA genome into messenger RNA (mRNA) and to replicate the genome. Viruses in this kingdom share a number of characteristics which promote rapid evolution, including high rates of genetic mutation, recombination, and reassortment.
ORF1ab refers collectively to two open reading frames (ORFs), ORF1a and ORF1b, that are conserved in the genomes of nidoviruses, a group of viruses that includes coronaviruses. The genes express large polyproteins that undergo proteolysis to form several nonstructural proteins with various functions in the viral life cycle, including proteases and the components of the replicase-transcriptase complex (RTC). Together the two ORFs are sometimes referred to as the replicase gene. They are related by a programmed ribosomal frameshift that allows the ribosome to continue translating past the stop codon at the end of ORF1a, in a -1 reading frame. The resulting polyproteins are known as pp1a and pp1ab.
Planarian secretory cell nidovirus (PSCNV) is a virus of the species Planidovirus 1, a nidovirus notable for its extremely large genome. At 41.1 kilobases, it is the largest known genome of an RNA virus. It was discovered by inspecting the transcriptomes of the planarian flatworm Schmidtea mediterranea and is the first known RNA virus infecting planarians. It was first described in 2018.















