
In cell biology, the nucleus is a membrane-bound organelle found in eukaryotic cells. Eukaryotes usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix, a network within the nucleus that adds mechanical support, much like the cytoskeleton supports the cell as a whole.

In cell biology, mitosis is a part of the cell cycle in which replicated chromosomes are separated into two new nuclei. Cell division gives rise to genetically identical cells in which the total number of chromosomes is maintained. Therefore, mitosis is also known as equational division. In general, mitosis is preceded by the S stage of interphase and is often followed by telophase and cytokinesis; which divides the cytoplasm, organelles and cell membrane of one cell into two new cells containing roughly equal shares of these cellular components. The different stages of Mitosis altogether define the mitotic (M) phase of an animal cell cycle—the division of the mother cell into two daughter cells genetically identical to each other.

Microtubules are polymers of tubulin that form part of the cytoskeleton and provide structure and shape to eukaryotic cells. Microtubules can grow as long as 50 micrometres and are highly dynamic. The outer diameter of a microtubule is between 23 and 27 nm while the inner diameter is between 11 and 15 nm. They are formed by the polymerization of a dimer of two globular proteins, alpha and beta tubulin into protofilaments that can then associate laterally to form a hollow tube, the microtubule. The most common form of a microtubule consists of 13 protofilaments in the tubular arrangement.

In cell biology, the spindle apparatus refers to the cytoskeletal structure of eukaryotic cells that forms during cell division to separate sister chromatids between daughter cells. It is referred to as the mitotic spindle during mitosis, a process that produces genetically identical daughter cells, or the meiotic spindle during meiosis, a process that produces gametes with half the number of chromosomes of the parent cell.
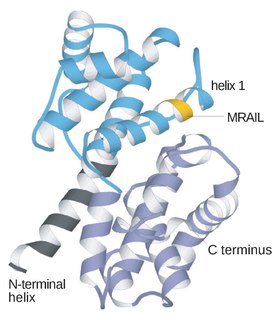
Cyclin is a family of proteins that controls the progression of a cell through the cell cycle by activating cyclin-dependent kinase (CDK) enzymes or group of enzymes required for synthesis of cell cycle.

The spindle checkpoint, also known as the metaphase-to-anaphase transition, the spindle assembly checkpoint (SAC), the metaphase checkpoint, or the mitotic checkpoint, is a cell cycle checkpoint during mitosis or meiosis that prevents the separation of the duplicated chromosomes (anaphase) until each chromosome is properly attached to the spindle. To achieve proper segregation, the two kinetochores on the sister chromatids must be attached to opposite spindle poles. Only this pattern of attachment will ensure that each daughter cell receives one copy of the chromosome. The defining biochemical feature of this checkpoint is the stimulation of the anaphase-promoting complex by M-phase cyclin-CDK complexes, which in turn causes the proteolytic destruction of cyclins and proteins that hold the sister chromatids together.

A kinetochore is a disc-shaped protein structure associated with duplicated chromatids in eukaryotic cells where the spindle fibers attach during cell division to pull sister chromatids apart. The kinetochore assembles on the centromere and links the chromosome to microtubule polymers from the mitotic spindle during mitosis and meiosis. The term kinetochore was first used in a footnote in a 1934 Cytology book by Lester W. Sharp and commonly accepted in 1936. Sharp's footnote reads: "The convenient term kinetochore has been suggested to the author by J. A. Moore", likely referring to John Alexander Moore who had joined Columbia University as a freshman in 1932.

A gene is a genetic unit of inheritance that consists of a segment of DNA that encodes for a functional RNA or a messenger RNA that is used to make proteins. All of the genes together make us the organism's genome and direct organismal phenotypes. Genes can be located in three distinct places in eukaryotic cells. The vast majority of genes are found in the chromosomes within the nucleus.
Jason Swedlow is an American-born cell biologist and light microscopist who is Professor of Quantitative Cell Biology at the School of Life Sciences, University of Dundee, Scotland. He is a co-founder of the Open Microscopy Environment and Glencoe Software. In 2021, he joined Wellcome Leap as a Program Director.

Aurora B kinase is a protein that functions in the attachment of the mitotic spindle to the centromere.

Cytoplasmic dynein 1 heavy chain 1 is a protein that in humans is encoded by the DYNC1H1 gene.

Interleukin enhancer-binding factor 2 is a protein that in humans is encoded by the ILF2 gene.

Kinesin-like protein KIF2C is a protein that in humans is encoded by the KIF2C gene.

Cytoplasmic linker associated protein 1, also known as CLASP1, is a protein which in humans is encoded by the CLASP1 gene.

Kinetochore protein Nuf2 is a protein that in humans is encoded by the NUF2 gene.

Centromere/kinetochore protein zw10 homolog is a protein that in humans is encoded by the ZW10 gene. This gene encodes a protein that is one of many involved in mechanisms to ensure proper chromosome segregation during cell division. The encoded protein binds to centromeres during the prophase, metaphase, and early anaphase cell division stages and to kinetochore microtubules during metaphase.
Iain Cheeseman investigates the role of the kinetochore, a group of proteins required for cell division and chromosome segregation. This core network of proteins facilitates the attachment of chromosomes to microtubule polymers—the spindle structures that attach to the ends of cells, pulling and dividing them during cell division. The kinetochore is critical to ensuring duplication without loss or damage to the genetic material. Cheeseman is also investigating the activities of the individual molecular machines that make up this structure and how these proteins are controlled and regulated.
The XMAP215/Dis1 family is a highly conserved group of microtubule-associated proteins (MAPs) in eukaryotic organisms. These proteins are unique MAPs because they primarily interact with the growing-end (plus-end) of microtubules. This special property classifies this protein family as plus-end tracking proteins (+TIPs).

Mary C. Dasso is an American biochemist known for research on chromosome segregation and the discovery of Ran GTPase. She is the acting scientific director of the division of intramural research and a senior investigator in the section on cell cycle regulation at the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
J. Richard McIntosh is a Distinguished Professor Emeritus in Molecular, Cellular, and Developmental Biology at the University of Colorado Boulder. McIntosh first graduated from Harvard with a BA in Physics in 1961, and again with a Ph.D. in Biophysics in 1968. He began his teaching career at Harvard but has spent most of his career at the University of Colorado Boulder. At the University of Colorado Boulder, McIntosh taught biology courses at both the undergraduate and graduate levels. Additionally, he created an undergraduate course in the biology of cancer towards the last several years of his teaching career. McIntosh's research career looks at a variety of things, including different parts of mitosis, microtubules, and motor proteins.
















