Definition
Ewens's sampling formula, introduced by Warren Ewens, states that under certain conditions (specified below), if a random sample of n gametes is taken from a population and classified according to the gene at a particular locus then the probability that there are a1 alleles represented once in the sample, and a2 alleles represented twice, and so on, is

for some positive number θ representing the population mutation rate, whenever  is a sequence of nonnegative integers such that
is a sequence of nonnegative integers such that

The phrase "under certain conditions" used above is made precise by the following assumptions:
- The sample size n is small by comparison to the size of the whole population; and
- The population is in statistical equilibrium under mutation and genetic drift and the role of selection at the locus in question is negligible; and
- Every mutant allele is novel.
This is a probability distribution on the set of all partitions of the integer n. Among probabilists and statisticians it is often called the multivariate Ewens distribution.

In statistics, an estimator is a rule for calculating an estimate of a given quantity based on observed data: thus the rule, the quantity of interest and its result are distinguished. For example, the sample mean is a commonly used estimator of the population mean.
The likelihood function is the joint probability mass of observed data viewed as a function of the parameters of a statistical model. Intuitively, the likelihood function is the probability of observing data assuming is the actual parameter.
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the frequency of an existing gene variant (allele) in a population due to random chance.
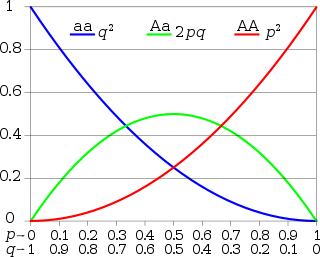
In population genetics, the Hardy–Weinberg principle, also known as the Hardy–Weinberg equilibrium, model, theorem, or law, states that allele and genotype frequencies in a population will remain constant from generation to generation in the absence of other evolutionary influences. These influences include genetic drift, mate choice, assortative mating, natural selection, sexual selection, mutation, gene flow, meiotic drive, genetic hitchhiking, population bottleneck, founder effect,inbreeding and outbreeding depression.
In probability and statistics, an exponential family is a parametric set of probability distributions of a certain form, specified below. This special form is chosen for mathematical convenience, including the enabling of the user to calculate expectations, covariances using differentiation based on some useful algebraic properties, as well as for generality, as exponential families are in a sense very natural sets of distributions to consider. The term exponential class is sometimes used in place of "exponential family", or the older term Koopman–Darmois family. Sometimes loosely referred to as "the" exponential family, this class of distributions is distinct because they all possess a variety of desirable properties, most importantly the existence of a sufficient statistic.
In statistics, Gibbs sampling or a Gibbs sampler is a Markov chain Monte Carlo (MCMC) algorithm for sampling from a specified multivariate probability distribution when direct sampling from the joint distribution is difficult, but sampling from the conditional distribution is more practical. This sequence can be used to approximate the joint distribution ; to approximate the marginal distribution of one of the variables, or some subset of the variables ; or to compute an integral. Typically, some of the variables correspond to observations whose values are known, and hence do not need to be sampled.

The unified neutral theory of biodiversity and biogeography is a theory and the title of a monograph by ecologist Stephen P. Hubbell. It aims to explain the diversity and relative abundance of species in ecological communities. Like other neutral theories of ecology, Hubbell assumes that the differences between members of an ecological community of trophically similar species are "neutral", or irrelevant to their success. This implies that niche differences do not influence abundance and the abundance of each species follows a random walk. The theory has sparked controversy, and some authors consider it a more complex version of other null models that fit the data better.
In statistics, the method of moments is a method of estimation of population parameters. The same principle is used to derive higher moments like skewness and kurtosis.

Genetic distance is a measure of the genetic divergence between species or between populations within a species, whether the distance measures time from common ancestor or degree of differentiation. Populations with many similar alleles have small genetic distances. This indicates that they are closely related and have a recent common ancestor.
Coalescent theory is a model of how alleles sampled from a population may have originated from a common ancestor. In the simplest case, coalescent theory assumes no recombination, no natural selection, and no gene flow or population structure, meaning that each variant is equally likely to have been passed from one generation to the next. The model looks backward in time, merging alleles into a single ancestral copy according to a random process in coalescence events. Under this model, the expected time between successive coalescence events increases almost exponentially back in time. Variance in the model comes from both the random passing of alleles from one generation to the next, and the random occurrence of mutations in these alleles.
In probability theory, the Chinese restaurant process is a discrete-time stochastic process, analogous to seating customers at tables in a restaurant. Imagine a restaurant with an infinite number of circular tables, each with infinite capacity. Customer 1 sits at the first table. The next customer either sits at the same table as customer 1, or the next table. This continues, with each customer choosing to either sit at an occupied table with a probability proportional to the number of customers already there, or an unoccupied table. At time n, the n customers have been partitioned among m ≤ n tables. The results of this process are exchangeable, meaning the order in which the customers sit does not affect the probability of the final distribution. This property greatly simplifies a number of problems in population genetics, linguistic analysis, and image recognition.
In natural language processing, latent Dirichlet allocation (LDA) is a Bayesian network for modeling automatically extracted topics in textual corpora. The LDA is an example of a Bayesian topic model. In this, observations are collected into documents, and each word's presence is attributable to one of the document's topics. Each document will contain a small number of topics.
In statistics, the multivariate t-distribution is a multivariate probability distribution. It is a generalization to random vectors of the Student's t-distribution, which is a distribution applicable to univariate random variables. While the case of a random matrix could be treated within this structure, the matrix t-distribution is distinct and makes particular use of the matrix structure.
In population genetics, the Watterson estimator is a method for describing the genetic diversity in a population. It was developed by Margaret Wu and G. A. Watterson in the 1970s. It is estimated by counting the number of polymorphic sites. It is a measure of the "population mutation rate" from the observed nucleotide diversity of a population. , where is the effective population size and is the per-generation mutation rate of the population of interest. The assumptions made are that there is a sample of haploid individuals from the population of interest, that there are infinitely many sites capable of varying, and that . Because the number of segregating sites counted will increase with the number of sequences looked at, the correction factor is used.
Tajima's D is a population genetic test statistic created by and named after the Japanese researcher Fumio Tajima. Tajima's D is computed as the difference between two measures of genetic diversity: the mean number of pairwise differences and the number of segregating sites, each scaled so that they are expected to be the same in a neutrally evolving population of constant size.
In statistics, a Pólya urn model, named after George Pólya, is a family of urn models that can be used to interpret many commonly used statistical models.
In population genetics, the allele frequency spectrum, sometimes called the site frequency spectrum, is the distribution of the allele frequencies of a given set of loci in a population or sample. Because an allele frequency spectrum is often a summary of or compared to sequenced samples of the whole population, it is a histogram with size depending on the number of sequenced individual chromosomes. Each entry in the frequency spectrum records the total number of loci with the corresponding derived allele frequency. Loci contributing to the frequency spectrum are assumed to be independently changing in frequency. Furthermore, loci are assumed to be biallelic, although extensions for multiallelic frequency spectra exist.
The Infinite sites model (ISM) is a mathematical model of molecular evolution first proposed by Motoo Kimura in 1969. Like other mutation models, the ISM provides a basis for understanding how mutation develops new alleles in DNA sequences. Using allele frequencies, it allows for the calculation of heterozygosity, or genetic diversity, in a finite population and for the estimation of genetic distances between populations of interest.
Multispecies Coalescent Process is a stochastic process model that describes the genealogical relationships for a sample of DNA sequences taken from several species. It represents the application of coalescent theory to the case of multiple species. The multispecies coalescent results in cases where the relationships among species for an individual gene can differ from the broader history of the species. It has important implications for the theory and practice of phylogenetics and for understanding genome evolution.
In probability theory, a branch of mathematics Poisson-Dirichlet distributions are probability distributions on the set of nonnegative, non-decreasing sequences with sum 1, depending on two parameters and . It can be defined as follows. One considers independent random variables such that follows the beta distribution of parameters and . Then, the Poisson-Dirichlet distribution of parameters and is the law of the random decreasing sequence containing and the products . This definition is due to Jim Pitman and Marc Yor. It generalizes Kingman's law, which corresponds to the particular case .






