
Leghemoglobin is an oxygen-carrying phytoglobin found in the nitrogen-fixing root nodules of leguminous plants. It is produced by these plants in response to the roots being colonized by nitrogen-fixing bacteria, termed rhizobia, as part of the symbiotic interaction between plant and bacterium: roots not colonized by Rhizobium do not synthesise leghemoglobin. Leghemoglobin has close chemical and structural similarities to hemoglobin, and, like hemoglobin, is red in colour. It was originally thought that the heme prosthetic group for plant leghemoglobin was provided by the bacterial symbiont within symbiotic root nodules. However, subsequent work shows that the plant host strongly expresses heme biosynthesis genes within nodules, and that activation of those genes correlates with leghemoglobin gene expression in developing nodules.
In biochemistry, an oxidoreductase is an enzyme that catalyzes the transfer of electrons from one molecule, the reductant, also called the electron donor, to another, the oxidant, also called the electron acceptor. This group of enzymes usually utilizes NADP+ or NAD+ as cofactors. Transmembrane oxidoreductases create electron transport chains in bacteria, chloroplasts and mitochondria, including respiratory complexes I, II and III. Some others can associate with biological membranes as peripheral membrane proteins or be anchored to the membranes through a single transmembrane helix.
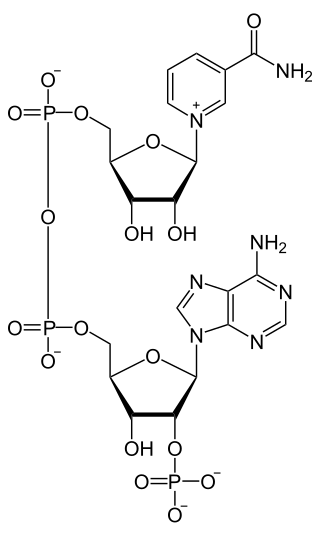
Nicotinamide adenine dinucleotide phosphate, abbreviated NADP+ or, in older notation, TPN (triphosphopyridine nucleotide), is a cofactor used in anabolic reactions, such as the Calvin cycle and lipid and nucleic acid syntheses, which require NADPH as a reducing agent ('hydrogen source'). NADPH is the reduced form of NADP+, the oxidized form. NADP+ is used by all forms of cellular life.
Microbial metabolism is the means by which a microbe obtains the energy and nutrients it needs to live and reproduce. Microbes use many different types of metabolic strategies and species can often be differentiated from each other based on metabolic characteristics. The specific metabolic properties of a microbe are the major factors in determining that microbe's ecological niche, and often allow for that microbe to be useful in industrial processes or responsible for biogeochemical cycles.

Dihydrolipoamide dehydrogenase (DLD), also known as dihydrolipoyl dehydrogenase, mitochondrial, is an enzyme that in humans is encoded by the DLD gene. DLD is a flavoprotein enzyme that oxidizes dihydrolipoamide to lipoamide.

Glyoxylate reductase, first isolated from spinach leaves, is an enzyme that catalyzes the reduction of glyoxylate to glycolate, using the cofactor NADH or NADPH.
In enzymology, a hydroxypyruvate reductase (EC 1.1.1.81) is an enzyme that catalyzes the chemical reaction
In enzymology, a mevaldate reductase (EC 1.1.1.32) is an enzyme that catalyzes the chemical reaction
In enzymology, a 4-hydroxybenzoate 3-monooxygenase [NAD(P)H] (EC 1.14.13.33) is an enzyme that catalyzes the chemical reaction
In enzymology, a 4-hydroxyphenylacetaldehyde oxime monooxygenase (EC 1.14.13.68) is an enzyme that catalyzes the chemical reaction
In enzymology, an anthraniloyl-CoA monooxygenase (EC 1.14.13.40) is an enzyme that catalyzes the chemical reaction
In enzymology, a ferredoxin–NAD+ reductase (EC 1.18.1.3) is an enzyme that catalyzes the chemical reaction:
In enzymology, a ferric-chelate reductase (EC 1.16.1.7) is an enzyme that catalyzes the chemical reaction
Flavin reductase a class of enzymes. There are a variety of flavin reductases, which bind free flavins and through hydrogen bonding, catalyze the reduction of these molecules to a reduced flavin. Riboflavin, or vitamin B, and flavin mononucleotide are two of the most well known flavins in the body and are used in a variety of processes which include metabolism of fat and ketones and the reduction of methemoglobin in erythrocytes. Flavin reductases are similar and often confused for ferric reductases because of their similar catalytic mechanism and structures.
In enzymology, a glutamate synthase (NADH) (EC 1.4.1.14) is an enzyme that catalyzes the chemical reaction
In enzymology, a hydroxylamine reductase (NADH) (EC 1.7.1.10) is an enzyme that catalyzes the chemical reaction.
In enzymology, a NAD(P)H dehydrogenase (quinone) (EC 1.6.5.2) is an enzyme that catalyzes the chemical reaction
In enzymology, a nitrite reductase [NAD(P)H] (EC 1.7.1.4) is an enzyme that catalyzes the chemical reaction
5-exo-hydroxycamphor dehydrogenase (EC 1.1.1.327, F-dehydrogenase, FdeH) is an enzyme with systematic name 5-exo-hydroxycamphor:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction

Phytoglobins are globular plant proteins classified into the globin superfamily, which contain a heme, i.e. protoporphyrin IX-Fe, prosthetic group. The earliest known phytoglobins are leghemoglobins, discovered in 1939 by Kubo after spectroscopic and chemical analysis of the red pigment of soybean root nodules. A few decades after Kubo's report the crystallization of a lupin phytoglobin by Vainshtein and collaborators revealed that the tertiary structure of this protein and that of the sperm whale myoglobin was remarkably similar, thus indicating that the phytoglobin discovered by Kubo did indeed correspond to a globin.









