Phosphopyruvate hydratase, usually known as enolase, is a metalloenzyme (EC 4.2.1.11) that catalyses the conversion of 2-phosphoglycerate (2-PG) to phosphoenolpyruvate (PEP), the ninth and penultimate step of glycolysis. The chemical reaction is:

Muconate lactonizing enzymes are involved in the breakdown of lignin-derived aromatics, catechol and protocatechuate, to citric acid cycle intermediates as a part of the β-ketoadipate pathway in soil microbes. Some bacterial species are also capable of dehalogenating chloroaromatic compounds by the action of chloromuconate lactonizing enzymes. MLEs consist of several strands which have variable reaction favorable parts therefore the configuration of the strands affect its ability to accept protons. The bacterial MLEs belong to the enolase superfamily, several structures from which are known. MLEs have an identifying structure made up of two proteins and two Magnesium ions as well as various classes depending on whether it is bacterial or eukaryotic. The reaction mechanism that MLEs undergo are the reverse of beta-elimination in which the enolate alpha-carbon is protonated. MLEs can undergo mutations caused by a deletion of catB structural genes which can cause some bacteria to lose its functions such as the ability to grow. Additional mutations to MLEs can cause its structure and function to alter and could cause the conformation to change therefore making it an inactive enzyme that is unable to bind its substrate. There is another enzyme called Mandelate Racemase that is very similar to MLEs in the structural way as well as them both being a part of the enolase superfamily. They both have the same end product even though they undergo different chemical reactions in order to reach the end product.
The enolase superfamily is a superfamily of enzymes, members of which catalyse a range of reactions.

2-Phosphoglyceric acid (2PG), or 2-phosphoglycerate, is a glyceric acid which serves as the substrate in the ninth step of glycolysis. It is catalyzed by enolase into phosphoenolpyruvate (PEP), the penultimate step in the conversion of glucose to pyruvate.
The lactate racemase enzyme (Lar) interconverts the D- and L-enantiomers of lactic acid. It is classified under the isomerase, racemase, epimerase, and enzyme acting on hydroxyl acids and derivatives classes of enzymes. It is found in certain halophilic archaea, such as Haloarcula marismortui, and in a few species of bacteria, such as several Lactobacillus species including Lactobacillus sakei, Lactobacillus curvatus, and Lactobacillus plantarum, as well as in non-lactic acid bacteria such as Clostridium beijerinckii. The gene encoding lactate racemase in L. plantarum was identified as larA and shown to be associated with a widespread maturation system involving larB, larC1, larC2, and larE. The optimal pH for its activity is 5.8-6.2 in L. sakei.

Methylmalonyl CoA epimerase is an enzyme involved in fatty acid catabolism that is encoded in human by the "MCEE" gene located on chromosome 2. It is routinely and incorrectly labeled as "methylmalonyl-CoA racemase". It is not a racemase because the CoA moiety has 5 other stereocenters.
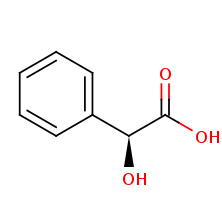
In enzymology, (S)-mandelate dehydrogenase (MDH), is an enzyme that catalyzes the chemical reaction.
In enzymology, a benzaldehyde dehydrogenase (NAD+) (EC 1.2.1.28) is an enzyme that catalyzes the chemical reaction
In enzymology, a benzaldehyde dehydrogenase (NADP+) (EC 1.2.1.7) is an enzyme that catalyzes the chemical reaction

In enzymology, an alanine racemase is an enzyme that catalyzes the chemical reaction

α-Methylacyl-CoA racemase is an enzyme that in humans is encoded by the AMACR gene. AMACR catalyzes the following chemical reaction:
In enzymology, an amino-acid racemase is an enzyme that catalyzes the chemical reaction
In enzymology, a methionine racemase is an enzyme that catalyzes the chemical reaction
In enzymology, a proline racemase is an enzyme that catalyzes the chemical reaction
In enzymology, a tartrate epimerase is an enzyme that catalyzes the chemical reaction
The enzyme benzoylformate decarboxylase (EC 4.1.1.7) catalyzes the following chemical reaction:

Gamma-enolase, also known as enolase 2 (ENO2) or neuron specific enolase (NSE), is an enzyme that in humans is encoded by the ENO2 gene. Gamma-enolase is a phosphopyruvate hydratase.
Enolase Deficiency is a rare genetic disorder of glucose metabolism. Partial deficiencies have been observed in several caucasian families. The deficiency is transmitted through an autosomal dominant inheritance pattern. The gene for Enolase 1 has been localized to Chromosome 1 in humans. Enolase deficiency, like other glycolytic enzyme deficiences, usually manifests in red blood cells as they rely entirely on anaerobic glycolysis. Enolase deficiency is associated with a spherocytic phenotype and can result in hemolytic anemia, which is responsible for the clinical signs of Enolase deficiency.

o-Succinylbenzoate synthase (OSBS) (EC 4.2.1.113) is an enzyme encoded by the menC gene in E.coli, and catalyzes the dehydration of 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate (SHCHC) to form 4-(2'-carboxyphenyl)-4-oxobutyrate, also called o-succinylbenzoate or OSB, hence the name of the enzyme. This reaction is the fourth step in the menaquinone biosynthetic pathway, which is used by bacteria to synthesize menaquinone, also known as vitamin K2.











