
A microRNA is a small single-stranded non-coding RNA molecule found in plants, animals and some viruses, that functions in RNA silencing and post-transcriptional regulation of gene expression. miRNAs function via base-pairing with complementary sequences within mRNA molecules. As a result, these mRNA molecules are silenced, by one or more of the following processes: (1) Cleavage of the mRNA strand into two pieces, (2) Destabilization of the mRNA through shortening of its poly(A) tail, and (3) Less efficient translation of the mRNA into proteins by ribosomes.
Shade avoidance is a set of responses that plants display when they are subjected to the shade of another plant. It often includes elongation, altered flowering time, increased apical dominance and altered partitioning of resources. This set of responses is collectively called the shade-avoidance syndrome (SAS).
In genetics and cell biology, repression is a mechanism often used to decrease or inhibit the expression of a gene. Removal of repression is called derepression. This mechanism may occur at different stages in the expression of a gene, with the result of increasing the overall RNA or protein products. Dysregulation of derepression mechanisms can result in altered gene expression patterns, which may lead to negative phenotypic consequences such as disease.

In molecular biology, mir-46 and mir-47 are microRNA expressed in C. elegans from related hairpin precursor sequences. The predicted hairpin precursor sequences for Drosophila mir-281 are also related and, hence, belong to this family. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequences are expressed from the 3' arms of the hairpin precursors.

MicroRNA (miRNA) precursor miR156 is a family of plant non-coding RNA. This microRNA has now been predicted or experimentally confirmed in a range of plant species. Animal miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. miR156 functions in the induction of flowering by suppressing the transcripts of SQUAMOSA-PROMOTER BINDING LIKE (SPL) transcription factors gene family. Its loading into ARGONAUTE5 is required for full functionality in Arabidopsis thaliana. In plants the precursor sequences may be longer, and the carpel factory (caf) enzyme appears to be involved in processing. In this case the mature sequence comes from the 5' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The products are thought to have regulatory roles through complementarity to mRNA.
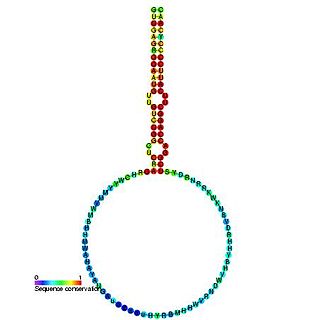
The plant mir-166 microRNA precursor is a small non-coding RNA gene. This microRNA (miRNA) has now been predicted or experimentally confirmed in a wide range of plant species. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 3' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The mature products are thought to have regulatory roles through complementarity to messenger RNA.

The mir-172 microRNA is thought to target mRNAs coding for APETALA2-like transcription factors. It has been verified experimentally in the model plant, Arabidopsis thaliana. The mature sequence is excised from the 3' arm of the hairpin.
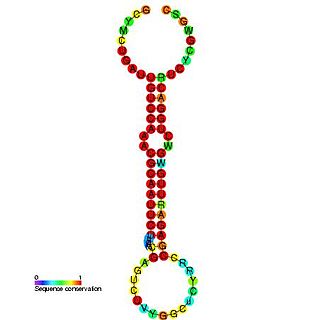
In molecular biology, the microRNA miR-219 was predicted in vertebrates by conservation between human, mouse and pufferfish and cloned in pufferfish. It was later predicted and confirmed experimentally in Drosophila. Homologs of miR-219 have since been predicted or experimentally confirmed in a wide range of species, including the platyhelminth Schmidtea mediterranea, several arthropod species and a wide range of vertebrates. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequence is excised from the 5' arm of the hairpin.

The mir-2 microRNA family includes the microRNA genes mir-2 and mir-13. Mir-2 is widespread in invertebrates, and it is the largest family of microRNAs in the model species Drosophila melanogaster. MicroRNAs from this family are produced from the 3' arm of the precursor hairpin. Leaman et al. showed that the miR-2 family regulates cell survival by translational repression of proapoptotic factors. Based on computational prediction of targets, a role in neural development and maintenance has been suggested.

mir-395 is a non-coding RNA called a microRNA that was identified in both Arabidopsis thaliana and Oryza sativa computationally and was later experimentally verified. mir-395 is thought to target mRNAs coding for ATP sulphurylases. The mature sequence is excised from the 3' arm of the hairpin.

mir-399 is a microRNA that was identified in both Arabidopsis thaliana and Oryza sativa computationally and was later experimentally verified. mir-399 is thought to target mRNAs coding for a phosphate transporter. The mature sequence is excised from the 3' arm of the hairpin. There are multiple copies of MIR399 in each plant genome, for example A. thaliana contains six microRNA precursors that all give rise to an almost identical mature miR-399 sequence.
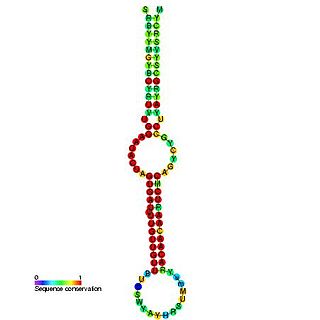
This family represents the microRNA (miRNA) precursor mir-7. This miRNA has been predicted or experimentally confirmed in a wide range of species. miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 5' arm of the precursor. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The involvement of Dicer in miRNA processing suggests a relationship with the phenomenon of RNA interference.
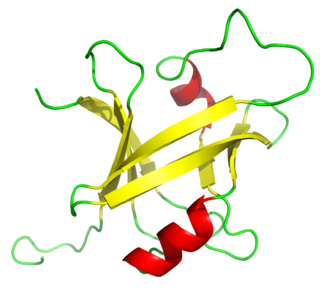
The B3 DNA binding domain (DBD) is a highly conserved domain found exclusively in transcription factors combined with other domains. It consists of 100-120 residues, includes seven beta strands and two alpha helices that form a DNA-binding pseudobarrel protein fold ; it interacts with the major groove of DNA.
Trans-acting siRNA are a class of small interfering RNA (siRNA) that repress gene expression through post-transcriptional gene silencing in land plants. Precursor transcripts from TAS loci are polyadenylated and converted to double-stranded RNA, and are then processed into 21-nucleotide-long RNA duplexes with overhangs. These segments are incorporated into a RNA-induced silencing complex (RISC) and direct the sequence-specific cleavage of target mRNA. Ta-siRNAs are classified as siRNA because they arise from double-stranded RNA (dsRNA).
In molecular biology mir-241 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-390 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-396 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-408 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-824 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.
In molecular biology mir-398 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.











