
The miR-103 microRNA precursor, is a short non-coding RNA gene involved in gene regulation. miR-103 and miR-107 have now been predicted or experimentally confirmed in human.
The miR-192 microRNA precursor, is a short non-coding RNA gene involved in gene regulation. miR-192 and miR-215 have now been predicted or experimentally confirmed in mouse and human.

In molecular biology, mir-46 and mir-47 are microRNA expressed in C. elegans from related hairpin precursor sequences. The predicted hairpin precursor sequences for Drosophila mir-281 are also related and, hence, belong to this family. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequences are expressed from the 3' arms of the hairpin precursors.

In molecular biology, miR-130 microRNA precursor is a small non-coding RNA that regulates gene expression. This microRNA has been identified in mouse, and in human. miR-130 appears to be vertebrate-specific miRNA and has now been predicted or experimentally confirmed in a range of vertebrate species. Mature microRNAs are processed from the precursor stem-loop by the Dicer enzyme. In this case, the mature sequence is excised from the 3' arm of the hairpin. It has been found that miR-130 is upregulated in a type of cancer called hepatocellular carcinoma. It has been shown that miR-130a is expressed in the hematopoietic stem/progenitor cell compartment but not in mature blood cells.

The miR-135 microRNA precursor is a small non-coding RNA that is involved in regulating gene expression. It has been shown to be expressed in human, mouse and rat. miR-135 has now been predicted or experimentally confirmed in a wide range of vertebrate species. Precursor microRNAs are ~70 nucleotides in length and are processed by the Dicer enzyme to produce the shorter 21-24 nucleotide mature sequence. In this case the mature sequence is excised from the 5' arm of the hairpin.

MicroRNA (miRNA) precursor miR156 is a family of plant non-coding RNA. This microRNA has now been predicted or experimentally confirmed in a range of plant species. Animal miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. miR156 functions in the induction of flowering by suppressing the transcripts of SQUAMOSA-PROMOTER BINDING LIKE (SPL) transcription factors gene family. Its loading into ARGONAUTE5 is required for full functionality in Arabidopsis thaliana. In plants the precursor sequences may be longer, and the carpel factory (caf) enzyme appears to be involved in processing. In this case the mature sequence comes from the 5' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The products are thought to have regulatory roles through complementarity to mRNA.
In molecular biology, mir-160 is a microRNA that has been predicted or experimentally confirmed in a range of plant species including Arabidopsis thaliana and Oryza sativa (rice). miR-160 is predicted to bind complementary sites in the untranslated regions of auxin response factor genes to regulate their expression. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequence is excised from the 5' arm of the hairpin.
The plant mir-166 microRNA precursor is a small non-coding RNA gene. This microRNA (miRNA) has now been predicted or experimentally confirmed in a wide range of plant species. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 3' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The mature products are thought to have regulatory roles through complementarity to messenger RNA.

The mir-172 microRNA is thought to target mRNAs coding for APETALA2-like transcription factors. It has been verified experimentally in the model plant, Arabidopsis thaliana. The mature sequence is excised from the 3' arm of the hairpin.
In molecular biology, miR-194 microRNA precursor is a small non-coding RNA gene that regulated gene expression. Its expression has been verified in mouse and in human. mir-194 appears to be a vertebrate-specific miRNA and has now been predicted or experimentally confirmed in a range of vertebrate species. The mature microRNA is processed from the longer hairpin precursor by the Dicer enzyme. In this case, the mature sequence is excised from the 5' arm of the hairpin.

The miR-29 microRNA precursor, or pre-miRNA, is a small RNA molecule in the shape of a stem-loop or hairpin. Each arm of the hairpin can be processed into one member of a closely related family of short non-coding RNAs that are involved in regulating gene expression. The processed, or "mature" products of the precursor molecule are known as microRNA (miRNA), and have been predicted or confirmed in a wide range of species.

The mir-2 microRNA family includes the microRNA genes mir-2 and mir-13. Mir-2 is widespread in invertebrates, and it is the largest family of microRNAs in the model species Drosophila melanogaster. MicroRNAs from this family are produced from the 3' arm of the precursor hairpin. Leaman et al. showed that the miR-2 family regulates cell survival by translational repression of proapoptotic factors. Based on computational prediction of targets, a role in neural development and maintenance has been suggested.

miR-30 microRNA precursor is a small non-coding RNA that regulates gene expression. Animal microRNAs are transcribed as pri-miRNA of varying length which in turns are processed in the nucleus by Drosha into ~70 nucleotide stem-loop precursor called pre-miRNA and subsequently processed by the Dicer enzyme to give a mature ~22 nucleotide product. In this case the mature sequence comes from both the 3' (miR-30) and 5' (mir-97-6) arms of the precursor. The products are thought to have regulatory roles through complementarity to mRNA.
The miR-34 microRNA precursor family are non-coding RNA molecules that, in mammals, give rise to three major mature miRNAs. The miR-34 family members were discovered computationally and later verified experimentally. The precursor miRNA stem-loop is processed in the cytoplasm of the cell, with the predominant miR-34 mature sequence excised from the 5' arm of the hairpin.

mir-395 is a non-coding RNA called a microRNA that was identified in both Arabidopsis thaliana and Oryza sativa computationally and was later experimentally verified. mir-395 is thought to target mRNAs coding for ATP sulphurylases. The mature sequence is excised from the 3' arm of the hairpin.
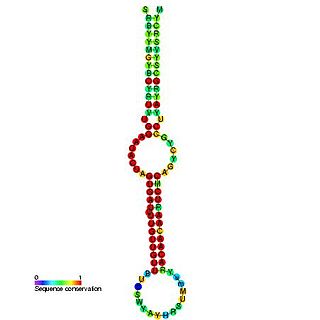
This family represents the microRNA (miRNA) precursor mir-7. This miRNA has been predicted or experimentally confirmed in a wide range of species. miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 5' arm of the precursor. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The involvement of Dicer in miRNA processing suggests a relationship with the phenomenon of RNA interference.
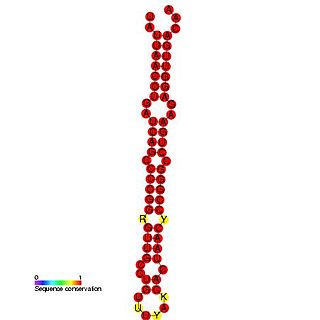
The mir-BHRF1-1 microRNA precursor found in Human herpesvirus 4 and Cercopithicine herpesvirus 15. In Epstein–Barr virus, mir-BHRF1-1 is found in the 5' UTR of the BHRF1 gene, which is known to encode a distant Bcl-2 homolog. The mature sequence is excised from the 5' arm of the hairpin. Two other miRNA precursors were found in this reading frame, namely Mir-BHRF1-2 and Mir-BHRF1-3.

The mir-BHRF1-3 microRNA precursor found in Human herpesvirus 4. In Epstein-Barr virus, mir-BHRF1-3 is found in the 3' UTR of the BHRF1 gene, which is known to encode a distant Bcl-2 homolog. The mature sequence is excised from the 5' arm of the hairpin. Two other miRNA precursors were found in this reading frame, namely Mir-BHRF1-1 and Mir-BHRF1-2.

miR-96 microRNA precursor is a small non-coding RNA that regulates gene expression. microRNAs are transcribed as ~80 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~23 nucleotide products. In this case the mature sequence comes from the 5′ arm of the precursor. The mature products are thought to have regulatory roles through complementarity to mRNA.
In molecular biology mir-396 microRNA is a short RNA molecule. MicroRNAs function to regulate the expression levels of other genes by several mechanisms.


















