
The miR-103 microRNA precursor, is a short non-coding RNA gene involved in gene regulation. miR-103 and miR-107 have now been predicted or experimentally confirmed in human.
In molecular biology, miR-148 is a microRNA whose expression has been demonstrated in human, mouse, rat and zebrafish. miR-148 has also been predicted in chicken.
The miR-192 microRNA precursor, is a short non-coding RNA gene involved in gene regulation. miR-192 and miR-215 have now been predicted or experimentally confirmed in mouse and human.

miR-101 microRNA precursor is a small non-coding RNA that regulates gene expression. Expression of miR-101 has been validated in both human and mouse. This microRNA appears to be specific to the vertebrates and has now been predicted or confirmed in a wide range of vertebrate species. The precursor microRNA is a stem-loop structure of about 70 nucleotides in length that is processed by the Dicer enzyme to form the 21-24 nucleotide mature microRNA. In this case the mature sequence is excised from the 3' arm of the hairpin.

The miR-10 microRNA precursor is a short non-coding RNA gene involved in gene regulation. It is part of an RNA gene family which contains miR-10, miR-51, miR-57, miR-99 and miR-100. miR-10, miR-99 and miR-100 have now been predicted or experimentally confirmed in a wide range of species. mir-51 and mir-57 have currently only been identified in the nematode Caenorhabditis elegans.

In molecular biology, miR-130 microRNA precursor is a small non-coding RNA that regulates gene expression. This microRNA has been identified in mouse, and in human. miR-130 appears to be vertebrate-specific miRNA and has now been predicted or experimentally confirmed in a range of vertebrate species. Mature microRNAs are processed from the precursor stem-loop by the Dicer enzyme. In this case, the mature sequence is excised from the 3' arm of the hairpin. It has been found that miR-130 is upregulated in a type of cancer called hepatocellular carcinoma. It has been shown that miR-130a is expressed in the hematopoietic stem/progenitor cell compartment but not in mature blood cells.

The miR-135 microRNA precursor is a small non-coding RNA that is involved in regulating gene expression. It has been shown to be expressed in human, mouse and rat. miR-135 has now been predicted or experimentally confirmed in a wide range of vertebrate species. Precursor microRNAs are ~70 nucleotides in length and are processed by the Dicer enzyme to produce the shorter 21-24 nucleotide mature sequence. In this case the mature sequence is excised from the 5' arm of the hairpin.

MicroRNA (miRNA) precursor miR156 is a family of plant non-coding RNA. This microRNA has now been predicted or experimentally confirmed in a range of plant species. Animal miRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In plants the precursor sequences may be longer, and the carpel factory (caf) enzyme appears to be involved in processing. In this case the mature sequence comes from the 5' arm of the precursor, and both Arabidopsis thaliana and rice genomes contain a number of related miRNA precursors which give rise to almost identical mature sequences. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The products are thought to have regulatory roles through complementarity to mRNA.
In molecular biology, mir-160 is a microRNA that has been predicted or experimentally confirmed in a range of plant species including Arabidopsis thaliana and Oryza sativa (rice). miR-160 is predicted to bind complementary sites in the untranslated regions of auxin response factor genes to regulate their expression. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequence is excised from the 5' arm of the hairpin.

miR-196 is a non-coding RNA called a microRNA that has been shown to be expressed in humans and mice. miR-196 appears to be a vertebrate specific microRNA and has now been predicted or experimentally confirmed in a wide range of vertebrate species. In many species the miRNA appears to be expressed from intergenic regions in HOX gene clusters. The hairpin precursors are predicted based on base pairing and cross-species conservation—their extents are not known. In this case the mature sequence is excised from the 5' arm of the hairpin.

The miR-199 microRNA precursor is a short non-coding RNA gene involved in gene regulation. miR-199 genes have now been predicted or experimentally confirmed in mouse, human and a further 21 other species. microRNAs are transcribed as ~70 nucleotide precursors and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. The mature products are thought to have regulatory roles through complementarity to mRNA.

There are 89 known sequences today in the microRNA 19 (miR-19) family but it will change quickly. They are found in a large number of vertebrate species. The miR-19 microRNA precursor is a small non-coding RNA molecule that regulates gene expression. Within the human and mouse genome there are three copies of this microRNA that are processed from multiple predicted precursor hairpins:

miR-218 microRNA precursor is a small non-coding RNA that regulates gene expression by antisense binding.
In molecular biology, the microRNA miR-219 was predicted in vertebrates by conservation between human, mouse and pufferfish and cloned in pufferfish. It was later predicted and confirmed experimentally in Drosophila. Homologs of miR-219 have since been predicted or experimentally confirmed in a wide range of species, including the platyhelminth Schmidtea mediterranea, several arthropod species and a wide range of vertebrates. The hairpin precursors are predicted based on base pairing and cross-species conservation; their extents are not known. In this case, the mature sequence is excised from the 5' arm of the hairpin.

The miR-29 microRNA precursor, or pre-miRNA, is a small RNA molecule in the shape of a stem-loop or hairpin. Each arm of the hairpin can be processed into one member of a closely related family of short non-coding RNAs that are involved in regulating gene expression. The processed, or "mature" products of the precursor molecule are known as microRNA (miRNA), and have been predicted or confirmed in a wide range of species.

The mir-2 microRNA family includes the microRNA genes mir-2 and mir-13. Mir-2 is widespread in invertebrates, and it is the largest family of microRNAs in the model species Drosophila melanogaster. MicroRNAs from this family are produced from the 3' arm of the precursor hairpin. Leaman et al. showed that the miR-2 family regulates cell survival by translational repression of proapoptotic factors. Based on computational prediction of targets, a role in neural development and maintenance has been suggested.
The miR-34 microRNA precursor family are non-coding RNA molecules that, in mammals, give rise to three major mature miRNAs. The miR-34 family members were discovered computationally and later verified experimentally. The precursor miRNA stem-loop is processed in the cytoplasm of the cell, with the predominant miR-34 mature sequence excised from the 5' arm of the hairpin.

The mir-6 microRNA precursor is a precursor microRNA specific to Drosophila species. In Drosophila melanogaster there are three mir-6 paralogs called dme-mir-6-1, dme-mir-6-2, dme-mir-6-3, which are clustered together in the genome. The extents of these hairpin precursors are estimated based on hairpin prediction. Each precursor is generated following the cleavage of a longer primary transcript in the nucleus, and is exported in the cytoplasm. In the cytoplasm, precursors are further processed by the enzyme Dicer, generating ~22 nucleotide products from each arm of the hairpin. The products generated from the 3' arm of each mir-6 precursor have identical sequences. Both 5' and 3' mature products are experimentally validated. Experimental data suggests that the mature products of mir-6 hairpins are expressed in the early embryo of Drosophila and target apoptotic genes such as hid, grim and rpr.

miR-338 is a family of brain-specific microRNA precursors found in mammals, including humans. The ~22 nucleotide mature miRNA sequence is excised from the precursor hairpin by the enzyme Dicer. This sequence then associates with RISC which effects RNA interference.
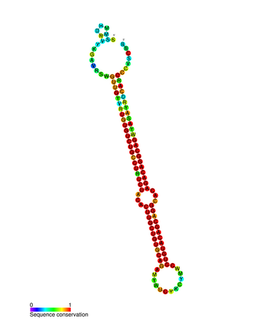
miR-214 is a vertebrate-specific family of microRNA precursors. The ~22 nucleotide mature miRNA sequence is excised from the precursor hairpin by the enzyme Dicer. This sequence then associates with RISC which effects RNA interference.



















