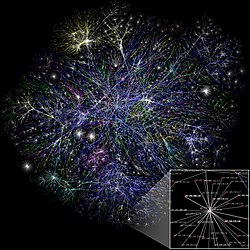
A scale-free network is a network whose degree distribution follows a power law, at least asymptotically. That is, the fraction P(k) of nodes in the network having k connections to other nodes goes for large values of k as

In mathematics, random graph is the general term to refer to probability distributions over graphs. Random graphs may be described simply by a probability distribution, or by a random process which generates them. The theory of random graphs lies at the intersection between graph theory and probability theory. From a mathematical perspective, random graphs are used to answer questions about the properties of typical graphs. Its practical applications are found in all areas in which complex networks need to be modeled – many random graph models are thus known, mirroring the diverse types of complex networks encountered in different areas. In a mathematical context, random graph refers almost exclusively to the Erdős–Rényi random graph model. In other contexts, any graph model may be referred to as a random graph.
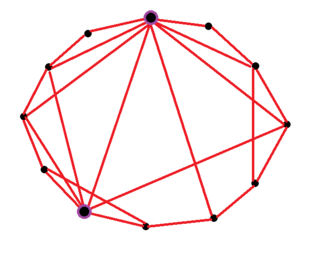
A small-world network is a mathematical graph in which most nodes are not neighbors of one another, but the neighbors of any given node are likely to be neighbors of each other. Due to this, most neighboring nodes can be reached from every other node by a small number of hops or steps. Specifically, a small-world network is defined to be a network where the typical distance L between two randomly chosen nodes grows proportionally to the logarithm of the number of nodes N in the network, that is:

In the study of graphs and networks, the degree of a node in a network is the number of connections it has to other nodes and the degree distribution is the probability distribution of these degrees over the whole network.

The Barabási–Albert (BA) model is an algorithm for generating random scale-free networks using a preferential attachment mechanism. Several natural and human-made systems, including the Internet, the World Wide Web, citation networks, and some social networks are thought to be approximately scale-free and certainly contain few nodes with unusually high degree as compared to the other nodes of the network. The BA model tries to explain the existence of such nodes in real networks. The algorithm is named for its inventors Albert-László Barabási and Réka Albert.

The Watts–Strogatz model is a random graph generation model that produces graphs with small-world properties, including short average path lengths and high clustering. It was proposed by Duncan J. Watts and Steven Strogatz in their article published in 1998 in the Nature scientific journal. The model also became known as the (Watts) beta model after Watts used to formulate it in his popular science book Six Degrees.
In complex network theory, the fitness model is a model of the evolution of a network: how the links between nodes change over time depends on the fitness of nodes. Fitter nodes attract more links at the expense of less fit nodes.
In the study of scale-free networks, a copying mechanism is a process by which such a network can form and grow, by means of repeated steps in which nodes are duplicated with mutations from existing nodes. Several variations have been studied. In the general copying model, a growing network starts as a small initial graph and, at each time step, a new vertex is added with a given number k of new outgoing edges. As a result of a stochastic selection, the neighbors of the new vertex are either chosen randomly among the existing vertices, or one existing vertex is randomly selected and k of its neighbors are "copied" as heads of the new edges.

In the mathematical field of graph theory, the Erdős–Rényi model refers to one of two closely related models for generating random graphs or the evolution of a random network. These models are named after Hungarian mathematicians Paul Erdős and Alfréd Rényi, who introduced one of the models in 1959. Edgar Gilbert introduced the other model contemporaneously with and independently of Erdős and Rényi. In the model of Erdős and Rényi, all graphs on a fixed vertex set with a fixed number of edges are equally likely. In the model introduced by Gilbert, also called the Erdős–Rényi–Gilbert model, each edge has a fixed probability of being present or absent, independently of the other edges. These models can be used in the probabilistic method to prove the existence of graphs satisfying various properties, or to provide a rigorous definition of what it means for a property to hold for almost all graphs.

Network science is an academic field which studies complex networks such as telecommunication networks, computer networks, biological networks, cognitive and semantic networks, and social networks, considering distinct elements or actors represented by nodes and the connections between the elements or actors as links. The field draws on theories and methods including graph theory from mathematics, statistical mechanics from physics, data mining and information visualization from computer science, inferential modeling from statistics, and social structure from sociology. The United States National Research Council defines network science as "the study of network representations of physical, biological, and social phenomena leading to predictive models of these phenomena."

Evolving networks are networks that change as a function of time. They are a natural extension of network science since almost all real world networks evolve over time, either by adding or removing nodes or links over time. Often all of these processes occur simultaneously, such as in social networks where people make and lose friends over time, thereby creating and destroying edges, and some people become part of new social networks or leave their networks, changing the nodes in the network. Evolving network concepts build on established network theory and are now being introduced into studying networks in many diverse fields.

Hierarchical network models are iterative algorithms for creating networks which are able to reproduce the unique properties of the scale-free topology and the high clustering of the nodes at the same time. These characteristics are widely observed in nature, from biology to language to some social networks.
Evolving networks are dynamic networks that change through time. In each period there are new nodes and edges that join the network while the old ones disappear. Such dynamic behaviour is characteristic for most real-world networks, regardless of their range - global or local. However, networks differ not only in their range but also in their topological structure. It is possible to distinguish:

Scientific collaboration network is a social network where nodes are scientists and links are co-authorships as the latter is one of the most well documented forms of scientific collaboration. It is an undirected, scale-free network where the degree distribution follows a power law with an exponential cutoff – most authors are sparsely connected while a few authors are intensively connected. The network has an assortative nature – hubs tend to link to other hubs and low-degree nodes tend to link to low-degree nodes. Assortativity is not structural, meaning that it is not a consequence of the degree distribution, but it is generated by some process that governs the network’s evolution.
In the context of complex networks, attack tolerance is the network's robustness meaning the ability to maintain the overall connectivity and diameter of the network as nodes are removed. Several graph metrics have been proposed to predicate network robustness. Algebraic connectivity is a graph metric that shows the best graph robustness among them.
Robustness, the ability to withstand failures and perturbations, is a critical attribute of many complex systems including complex networks.
In network science, preferential attachment means that nodes of a network tend to connect to those nodes which have more links. If the network is growing and new nodes tend to connect to existing ones with linear probability in the degree of the existing nodes then preferential attachment leads to a scale-free network. If this probability is sub-linear then the network’s degree distribution is stretched exponential and hubs are much smaller than in a scale-free network. If this probability is super-linear then almost all nodes are connected to a few hubs. According to Kunegis, Blattner, and Moser several online networks follow a non-linear preferential attachment model. Communication networks and online contact networks are sub-linear while interaction networks are super-linear. The co-author network among scientists also shows the signs of sub-linear preferential attachment.

The Bianconi–Barabási model is a model in network science that explains the growth of complex evolving networks. This model can explain that nodes with different characteristics acquire links at different rates. It predicts that a node's growth depends on its fitness and can calculate the degree distribution. The Bianconi–Barabási model is named after its inventors Ginestra Bianconi and Albert-László Barabási. This model is a variant of the Barabási–Albert model. The model can be mapped to a Bose gas and this mapping can predict a topological phase transition between a "rich-get-richer" phase and a "winner-takes-all" phase.

In network science, a hub is a node with a number of links that greatly exceeds the average. Emergence of hubs is a consequence of a scale-free property of networks. While hubs cannot be observed in a random network, they are expected to emerge in scale-free networks. The uprise of hubs in scale-free networks is associated with power-law distribution. Hubs have a significant impact on the network topology. Hubs can be found in many real networks, such as the brain or the Internet.

In the scale-free network theory, a mediation-driven attachment (MDA) model appears to embody a preferential attachment rule tacitly rather than explicitly. According to MDA rule, a new node first picks a node from the existing network at random and connect itself not with that but with one of the neighbors also picked at random.