
Coenzyme A (CoA, SHCoA, CoASH) is a coenzyme, notable for its role in the synthesis and oxidation of fatty acids, and the oxidation of pyruvate in the citric acid cycle. All genomes sequenced to date encode enzymes that use coenzyme A as a substrate, and around 4% of cellular enzymes use it (or a thioester) as a substrate. In humans, CoA biosynthesis requires cysteine, pantothenate (vitamin B5), and adenosine triphosphate (ATP).

Malonyl-CoA is a coenzyme A derivative of malonic acid.

The acyl carrier protein (ACP) is a cofactor of both fatty acid and polyketide biosynthesis machinery. It is one of the most abundant proteins in cells of E. coli. In both cases, the growing chain is bound to the ACP via a thioester derived from the distal thiol of a 4'-phosphopantetheine moiety.
In biochemistry, fatty acid synthesis is the creation of fatty acids from acetyl-CoA and NADPH through the action of enzymes called fatty acid synthases. This process takes place in the cytoplasm of the cell. Most of the acetyl-CoA which is converted into fatty acids is derived from carbohydrates via the glycolytic pathway. The glycolytic pathway also provides the glycerol with which three fatty acids can combine to form triglycerides, the final product of the lipogenic process. When only two fatty acids combine with glycerol and the third alcohol group is phosphorylated with a group such as phosphatidylcholine, a phospholipid is formed. Phospholipids form the bulk of the lipid bilayers that make up cell membranes and surrounds the organelles within the cells. In addition to cytosolic fatty acid synthesis, there is also mitochondrial fatty acid synthesis (mtFASII), in which malonyl-CoA is formed from malonic acid with the help of malonyl-CoA synthetase (ACSF3), which then becomes the final product octanoyl-ACP (C8) via further intermediate steps.

In molecular biology, Beta-ketoacyl-ACP synthase EC 2.3.1.41, is an enzyme involved in fatty acid synthesis. It typically uses malonyl-CoA as a carbon source to elongate ACP-bound acyl species, resulting in the formation of ACP-bound β-ketoacyl species such as acetoacetyl-ACP.
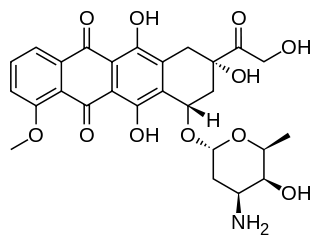
Doxorubicin (DXR) is a 14-hydroxylated version of daunorubicin, the immediate precursor of DXR in its biosynthetic pathway. Daunorubicin is more abundantly found as a natural product because it is produced by a number of different wild type strains of streptomyces. In contrast, only one known non-wild type species, streptomyces peucetius subspecies caesius ATCC 27952, was initially found to be capable of producing the more widely used doxorubicin. This strain was created by Arcamone et al. in 1969 by mutating a strain producing daunorubicin, but not DXR, at least in detectable quantities. Subsequently, Hutchinson's group showed that under special environmental conditions, or by the introduction of genetic modifications, other strains of streptomyces can produce doxorubicin. His group has also cloned many of the genes required for DXR production, although not all of them have been fully characterized. In 1996, Strohl's group discovered, isolated and characterized dox A, the gene encoding the enzyme that converts daunorubicin into DXR. By 1999, they produced recombinant Dox A, a Cytochrome P450 oxidase, and found that it catalyzes multiple steps in DXR biosynthesis, including steps leading to daunorubicin. This was significant because it became clear that all daunorubicin producing strains have the necessary genes to produce DXR, the much more therapeutically important of the two. Hutchinson's group went on to develop methods to improve the yield of DXR, from the fermentation process used in its commercial production, not only by introducing Dox A encoding plasmids, but also by introducing mutations to deactivate enzymes that shunt DXR precursors to less useful products, for example baumycin-like glycosides. Some triple mutants, that also over-expressed Dox A, were able to double the yield of DXR. This is of more than academic interest because at that time DXR cost about $1.37 million per kg and current production in 1999 was 225 kg per annum. More efficient production techniques have brought the price down to $1.1 million per kg for the non-liposomal formulation. Although DXR can be produced semi-synthetically from daunorubicin, the process involves electrophilic bromination and multiple steps and the yield is poor. Since daunorubicin is produced by fermentation, it would be ideal if the bacteria could complete DXR synthesis more effectively.
The enzyme [acyl-carrier-protein] phosphodiesterase (EC 3.1.4.14) catalyzes the reaction
In enzymology, a [acyl-carrier-protein] S-acetyltransferase is an enzyme that catalyzes the reversible chemical reaction
In enzymology, a beta-ketoacyl-acyl-carrier-protein synthase I is an enzyme that catalyzes the chemical reaction
In enzymology, a beta-ketoacyl-acyl-carrier-protein synthase II (EC 2.3.1.179) is an enzyme that catalyzes the chemical reaction

In enzymology, a β-ketoacyl-[acyl-carrier-protein] synthase III (EC 2.3.1.180) is an enzyme that catalyzes the chemical reaction
In enzymology, an erythronolide synthase is an enzyme that catalyzes the chemical reaction

Fatty-acyl-CoA Synthase, or more commonly known as yeast fatty acid synthase, is an enzyme complex responsible for fatty acid biosynthesis, and is of Type I Fatty Acid Synthesis (FAS). Yeast fatty acid synthase plays a pivotal role in fatty acid synthesis. It is a 2.6 MDa barrel shaped complex and is composed of two, unique multi-functional subunits: alpha and beta. Together, the alpha and beta units are arranged in an α6β6 structure. The catalytic activities of this enzyme complex involves a coordination system of enzymatic reactions between the alpha and beta subunits. The enzyme complex therefore consists of six functional centers for fatty acid synthesis.

In enzymology and molecular biology, a holo-[acyl-carrier-protein] synthase is an enzyme that catalyzes the chemical reaction:

Malonyl CoA-acyl carrier protein transacylase, mitochondrial is an enzyme that in humans is encoded by the MCAT gene.

Germicidins are a groups of natural products arising from Streptomyces species that acts as autoregulatory inhibitor of spore germination. In Streptomyces viriochromogenes, low concentrations inhibit germination of its own arthrospores, and higher concentrations inhibit porcine Na+/K+ -activated ATPase. Inhibitory effects on germination are also observed when germicidin from Streptomyces is applied to Lepidium sativum. Germicidins and other natural products present potential use as pharmaceuticals, and in this case, those with possible antibiotic or antifungal activity.
Very-long-chain 3-oxoacyl-CoA synthase (EC 2.3.1.199, very-long-chain 3-ketoacyl-CoA synthase, very-long-chain beta-ketoacyl-CoA synthase, condensing enzyme, CUT1, CER6, FAE1, KCS, ELO) is an enzyme with systematic name malonyl-CoA:very-long-chain acyl-CoA malonyltransferase (decarboxylating and thioester-hydrolysing). This enzyme catalyses the following chemical reaction
3-hydroxydecanoyl-(acyl-carrier-protein) dehydratase (EC 4.2.1.60, D-3-hydroxydecanoyl-[acyl-carrier protein] dehydratase, 3-hydroxydecanoyl-acyl carrier protein dehydrase, 3-hydroxydecanoyl-acyl carrier protein dehydratase, β-hydroxydecanoyl thioester dehydrase, β-hydroxydecanoate dehydrase, beta-hydroxydecanoyl thiol ester dehydrase, FabA, β-hydroxyacyl-acyl carrier protein dehydratase, HDDase, β-hydroxyacyl-ACP dehydrase, (3R)-3-hydroxydecanoyl-[acyl-carrier-protein] hydro-lyase) is an enzyme with systematic name (3R)-3-hydroxydecanoyl-(acyl-carrier protein) hydro-lyase. This enzyme catalyses the following chemical reaction

Ketoacyl synthases (KSs) catalyze the condensation reaction of acyl-CoA or acyl-acyl ACP with malonyl-CoA to form 3-ketoacyl-CoA or with malonyl-ACP to form 3-ketoacyl-ACP. This reaction is a key step in the fatty acid synthesis cycle, as the resulting acyl chain is two carbon atoms longer than before. KSs exist as individual enzymes, as they do in type II fatty acid synthesis and type II polyketide synthesis, or as domains in large multidomain enzymes, such as type I fatty acid synthases (FASs) and polyketide synthases (PKSs). KSs are divided into five families: KS1, KS2, KS3, KS4, and KS5.
Elongase is a generic term for an enzyme that extends the length of fatty acid. The nomenclature is not applied rigorously. Often, elongase refers to enzymes that produce very long chain fatty acids. Sometimes, elongase also includes unsaturases, which introduce C=C double bonds in the backbone. Because fatty acids and their derivatives are biochemically influential, elongases are of considerable interest.












