
Biological carbon fixation or сarbon assimilation is the process by which inorganic carbon is converted to organic compounds by living organisms. The compounds are then used to store energy and as structure for other biomolecules. Carbon is primarily fixed through photosynthesis, but some organisms use a process called chemosynthesis in the absence of sunlight.
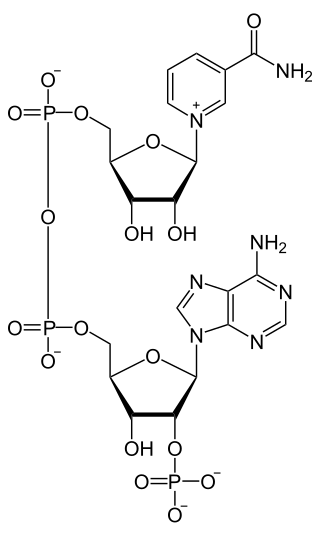
Nicotinamide adenine dinucleotide phosphate, abbreviated NADP+ or, in older notation, TPN (triphosphopyridine nucleotide), is a cofactor used in anabolic reactions, such as the Calvin cycle and lipid and nucleic acid syntheses, which require NADPH as a reducing agent ('hydrogen source'). NADPH is the reduced form of NADP+, the oxidized form. NADP+ is used by all forms of cellular life.

In enzymology, aldose reductase is a cytosolic NADPH-dependent oxidoreductase that catalyzes the reduction of a variety of aldehydes and carbonyls, including monosaccharides. It is primarily known for catalyzing the reduction of glucose to sorbitol, the first step in polyol pathway of glucose metabolism.

Nitrosopumilus maritimus is an extremely common archaeon living in seawater. It is the first member of the Group 1a Nitrososphaerota to be isolated in pure culture. Gene sequences suggest that the Group 1a Nitrososphaerota are ubiquitous with the oligotrophic surface ocean and can be found in most non-coastal marine waters around the planet. It is one of the smallest living organisms at 0.2 micrometers in diameter. Cells in the species N. maritimus are shaped like peanuts and can be found both as individuals and in loose aggregates. They oxidize ammonia to nitrite and members of N. maritimus can oxidize ammonia at levels as low as 10 nanomolar, near the limit to sustain its life. Archaea in the species N. maritimus live in oxygen-depleted habitats. Oxygen needed for ammonia oxidation might be produced by novel pathway which generates oxygen and dinitrogen. N. maritimus is thus among organisms which are able to produce oxygen in dark.
In enzymology, an anthraniloyl-CoA monooxygenase (EC 1.14.13.40) is an enzyme that catalyzes the chemical reaction
In enzymology, a ferredoxin-NADP+ reductase (EC 1.18.1.2) abbreviated FNR, is an enzyme that catalyzes the chemical reaction

[Methionine synthase] reductase, or Methionine synthase reductase, encoded by the gene MTRR, is an enzyme that is responsible for the reduction of methionine synthase inside human body. This enzyme is crucial for maintaining the one carbon metabolism, specifically the folate cycle. The enzyme employs one coenzyme, flavoprotein.
In enzymology, a CoA-glutathione reductase (EC 1.8.1.10) is an enzyme that catalyzes the chemical reaction
The 3-hydroxypropionate bicycle, also known as the 3-hydroxypropionate pathway, is a process that allows some bacteria to generate 3-hydroxypropionate usingcarbon dioxide. In this pathway CO2 is fixed (i.e. incorporated) by the action of two enzymes, acetyl-CoA carboxylase and propionyl-CoA carboxylase. These enzymes generate malonyl-CoA and (S)-methylmalonyl-CoA, respectively. Malonyl-CoA, in a series of reactions, is further split into acetyl-CoA and glyoxylate. Glyoxylate is incorporated into beta-methylmalyl-coA which is then split, again through a series of reactions, to release pyruvate as well as acetate, which is used to replenish the cycle. This pathway has been demonstrated in Chloroflexus, a nonsulfur photosynthetic bacterium; however, other studies suggest that 3-hydroxypropionate bicycle is used by several chemotrophic archaea.

In molecular biology, the HMG-CoA reductase family is a family of enzymes which participate in the mevalonate pathway, the metabolic pathway that produces cholesterol and other isoprenoids.
3-hydroxypropionate dehydrogenase (NADP+) (EC 1.1.1.298) is an enzyme with systematic name 3-hydroxypropionate:NADP+ oxidoreductase. This enzyme catalyses the following chemical reaction
Malonyl CoA reductase (malonate semialdehyde-forming) (EC 1.2.1.75, NADP-dependent malonyl CoA reductase, malonyl CoA reductase (NADP)) is an enzyme with systematic name malonate semialdehyde:NADP+ oxidoreductase (malonate semialdehyde-forming). This enzyme catalyse the following chemical reaction
Succinate-semialdehyde dehydrogenase (acylating) (EC 1.2.1.76, succinyl-coA reductase, coenzyme-A-dependent succinate-semialdehyde dehydrogenase) is an enzyme with systematic name succinate semialdehyde:NADP+ oxidoreductase (CoA-acylating). This enzyme catalyses the following chemical reaction
3,4-Dehydroadipyl-CoA semialdehyde dehydrogenase (NADP+) (EC 1.2.1.77, BoxD, 3,4-dehydroadipyl-CoA semialdehyde dehydrogenase) is an enzyme with systematic name 3,4-didehydroadipyl-CoA semialdehyde:NADP+ oxidoreductase. This enzyme catalyses the following chemical reaction

Crotonyl-CoA carboxylase/reductase (EC 1.3.1.85, CCR, crotonyl-CoA reductase (carboxylating)) is an enzyme with systematic name (2S)-ethylmalonyl-CoA:NADP+ oxidoreductase (decarboxylating). This enzyme catalyses the following chemical reaction
Crotonyl-CoA reductase (EC 1.3.1.86, butyryl-CoA dehydrogenase, butyryl dehydrogenase, unsaturated acyl-CoA reductase, ethylene reductase, enoyl-coenzyme A reductase, unsaturated acyl coenzyme A reductase, butyryl coenzyme A dehydrogenase, short-chain acyl CoA dehydrogenase, short-chain acyl-coenzyme A dehydrogenase, 3-hydroxyacyl CoA reductase, butanoyl-CoA:(acceptor) 2,3-oxidoreductase, CCR) is an enzyme with systematic name butanoyl-CoA:NADP+ 2,3-oxidoreductase. This enzyme catalyses the following chemical reaction
Acrylyl-CoA reductase (NADH) (EC 1.3.1.95) is an enzyme with systematic name propanoyl-CoA:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
3-hydroxypropionyl-CoA dehydratase (EC 4.2.1.116) is an enzyme with systematic name 3-hydroxypropionyl-CoA hydro-lyase. This enzyme catalyses the following chemical reaction
4-hydroxybutanoyl-CoA dehydratase (EC 4.2.1.120) is an enzyme with systematic name 4-hydroxybutanoyl-CoA hydro-lyase. This enzyme catalyses the following chemical reaction
3-Hydroxypropionyl-CoA synthase is an enzyme with systematic name hydroxypropionate:CoA ligase (AMP-forming). This enzyme catalyses the following chemical reaction







