A restriction enzyme, restriction endonuclease, REase, ENase orrestrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction

In biochemistry, the DNA methyltransferase family of enzymes catalyze the transfer of a methyl group to DNA. DNA methylation serves a wide variety of biological functions. All the known DNA methyltransferases use S-adenosyl methionine (SAM) as the methyl donor.

In biochemistry, a nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.

DnaA is a protein that activates initiation of DNA replication in bacteria. Based on the Replicon Model, a positively active initiator molecule contacts with a particular spot on a circular chromosome called the replicator to start DNA replication. It is a replication initiation factor which promotes the unwinding of DNA at oriC. The DnaA proteins found in all bacteria engage with the DnaA boxes to start chromosomal replication. In addition to the DnaA protein, its concentration, binding to DnaA-boxes, and binding of ATP or ADP, we will cover the regulation of the DnaA gene, the unique characteristics of the DnaA gene expression, promoter strength, and translation efficiency. The onset of the initiation phase of DNA replication is determined by the concentration of DnaA. DnaA accumulates during growth and then triggers the initiation of replication. Replication begins with active DnaA binding to 9-mer (9-bp) repeats upstream of oriC. Binding of DnaA leads to strand separation at the 13-mer repeats. This binding causes the DNA to loop in preparation for melting open by the helicase DnaB.
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. It is the only known enzyme to actively contribute negative supercoiling to DNA, while it also is capable of relaxing positive supercoils. It does so by looping the template to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite Plasmodium falciparum and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiotics, including nalidixic acid, novobiocin, albicidin, and ciprofloxacin.

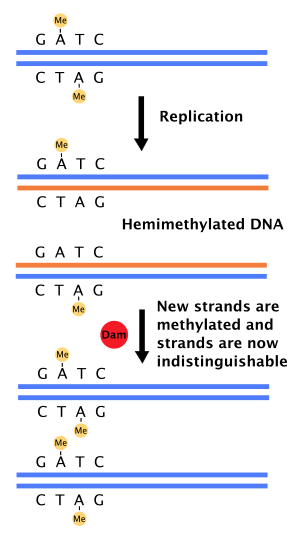
DNA methylation is a biological process by which methyl groups are added to the DNA molecule. Methylation can change the activity of a DNA segment without changing the sequence. When located in a gene promoter, DNA methylation typically acts to repress gene transcription. In mammals, DNA methylation is essential for normal development and is associated with a number of key processes including genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging, and carcinogenesis.
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically, while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.
P1 is a temperate bacteriophage that infects Escherichia coli and some other bacteria. When undergoing a lysogenic cycle the phage genome exists as a plasmid in the bacterium unlike other phages that integrate into the host DNA. P1 has an icosahedral head containing the DNA attached to a contractile tail with six tail fibers. The P1 phage has gained research interest because it can be used to transfer DNA from one bacterial cell to another in a process known as transduction. As it replicates during its lytic cycle it captures fragments of the host chromosome. If the resulting viral particles are used to infect a different host the captured DNA fragments can be integrated into the new host's genome. This method of in vivo genetic engineering was widely used for many years and is still used today, though to a lesser extent. P1 can also be used to create the P1-derived artificial chromosome cloning vector which can carry relatively large fragments of DNA. P1 encodes a site-specific recombinase, Cre, that is widely used to carry out cell-specific or time-specific DNA recombination by flanking the target DNA with loxP sites.
Prokaryotic DNA Replication is the process by which a prokaryote duplicates its DNA into another copy that is passed on to daughter cells. Although it is often studied in the model organism E. coli, other bacteria show many similarities. Replication is bi-directional and originates at a single origin of replication (OriC). It consists of three steps: Initiation, elongation, and termination.
Deoxyribonuclease IV (phage-T4-induced) is catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.

In enzymology, a protein-glutamate O-methyltransferase is an enzyme that catalyzes the chemical reaction
In biology, phase variation is a method for dealing with rapidly varying environments without requiring random mutation. It involves the variation of protein expression, frequently in an on-off fashion, within different parts of a bacterial population. As such the phenotype can switch at frequencies that are much higher than classical mutation rates. Phase variation contributes to virulence by generating heterogeneity. Although it has been most commonly studied in the context of immune evasion, it is observed in many other areas as well and is employed by various types of bacteria, including Salmonella species.
DNA adenine methyltransferase identification, often abbreviated DamID, is a molecular biology protocol used to map the binding sites of DNA- and chromatin-binding proteins in eukaryotes. DamID identifies binding sites by expressing the proposed DNA-binding protein as a fusion protein with DNA methyltransferase. Binding of the protein of interest to DNA localizes the methyltransferase in the region of the binding site. Adenine methylation does not occur naturally in eukaryotes and therefore adenine methylation in any region can be concluded to have been caused by the fusion protein, implying the region is located near a binding site. DamID is an alternate method to ChIP-on-chip or ChIP-seq.

A circular chromosome is a chromosome in bacteria, archaea, mitochondria, and chloroplasts, in the form of a molecule of circular DNA, unlike the linear chromosome of most eukaryotes.

In molecular biology the SeqA protein is found in bacteria and archaea. The function of this protein is highly important in DNA replication. The protein negatively regulates the initiation of DNA replication at the origin of replication, in Escherichia coli, OriC. Additionally the protein plays a further role in sequestration. The importance of this protein is vital, without its help in DNA replication, cell division and other crucial processes could not occur. This protein domain is thought to be part of a much larger protein complex which includes other proteins such as SeqB.
16S rRNA (adenine1518-N6/adenine1519-N6)-dimethyltransferase (EC 2.1.1.182, S-adenosylmethionine-6-N',N'-adenosyl (rRNA) dimethyltransferase, KsgA, ksgA methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:16S rRNA (adenine1518-N6/adenine1519-N6)-dimethyltransferase. This enzyme catalyses the following chemical reaction
23S rRNA (guanine745-N1)-methyltransferase (EC 2.1.1.187, Rlma(I), Rlma1, 23S rRNA m1G745 methyltransferase, YebH, RlmAI methyltransferase, ribosomal RNA(m1G)-methylase, rRNA(m1G)methylase, RrmA, 23S rRNA:m1G745 methyltransferase) is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (guanine745-N1)-methyltransferase. This enzyme catalyses the following chemical reaction
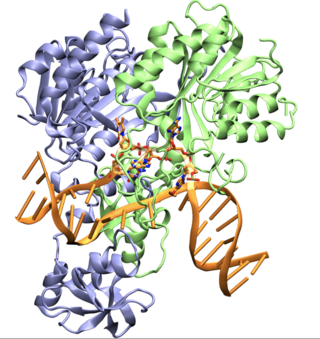
CcrM is an orphan DNA methyltransferase, that is involved in controlling gene expression in most Alphaproteobacteria. This enzyme modifies DNA by catalyzing the transference of a methyl group from the S-adenosyl-L methionine substrate to the N6 position of an adenine base in the sequence 5'-GANTC-3' with high specificity. In some lineages such as SAR11, the homologous enzymes possess 5'-GAWTC-3' specificity. In Caulobacter crescentus Ccrm is produced at the end of the replication cycle when Ccrm recognition sites are hemimethylated, rapidly methylating the DNA. CcrM is essential in other Alphaproteobacteria but is role is not yet determined. CcrM is a highly specific methyltransferase with a novel DNA recognition mechanism.