
Enzyme kinetics is the study of the rates of enzyme-catalysed chemical reactions. In enzyme kinetics, the reaction rate is measured and the effects of varying the conditions of the reaction are investigated. Studying an enzyme's kinetics in this way can reveal the catalytic mechanism of this enzyme, its role in metabolism, how its activity is controlled, and how a drug or a modifier might affect the rate.

The enzyme argininosuccinate lyase (EC 4.3.2.1, ASL, argininosuccinase; systematic name 2-(N ω-L-arginino)succinate arginine-lyase (fumarate-forming)) catalyzes the reversible breakdown of argininosuccinate:
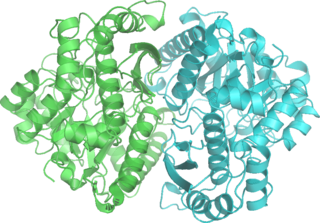
Phosphopyruvate hydratase, usually known as enolase, is a metalloenzyme (EC 4.2.1.11) that catalyses the conversion of 2-phosphoglycerate (2-PG) to phosphoenolpyruvate (PEP), the ninth and penultimate step of glycolysis. The chemical reaction is:

Orotidine 5'-phosphate decarboxylase or orotidylate decarboxylase is an enzyme involved in pyrimidine biosynthesis. It catalyzes the decarboxylation of orotidine monophosphate (OMP) to form uridine monophosphate (UMP). The function of this enzyme is essential to the de novo biosynthesis of the pyrimidine nucleotides uridine triphosphate, cytidine triphosphate, and thymidine triphosphate. OMP decarboxylase has been a frequent target for scientific investigation because of its demonstrated extreme catalytic efficiency and its usefulness as a selection marker for yeast strain engineering.
Transition state analogs, are chemical compounds with a chemical structure that resembles the transition state of a substrate molecule in an enzyme-catalyzed chemical reaction. Enzymes interact with a substrate by means of strain or distortions, moving the substrate towards the transition state. Transition state analogs can be used as inhibitors in enzyme-catalyzed reactions by blocking the active site of the enzyme. Theory suggests that enzyme inhibitors which resembled the transition state structure would bind more tightly to the enzyme than the actual substrate. Examples of drugs that are transition state analog inhibitors include flu medications such as the neuraminidase inhibitor oseltamivir and the HIV protease inhibitors saquinavir in the treatment of AIDS.
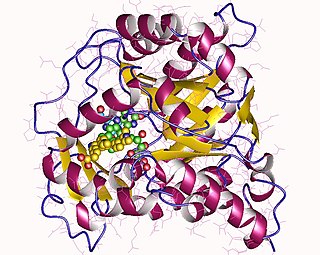
Dihydroorotate dehydrogenase (DHODH) is an enzyme that in humans is encoded by the DHODH gene on chromosome 16. The protein encoded by this gene catalyzes the fourth enzymatic step, the ubiquinone-mediated oxidation of dihydroorotate to orotate, in de novo pyrimidine biosynthesis. This protein is a mitochondrial protein located on the outer surface of the inner mitochondrial membrane (IMM). Inhibitors of this enzyme are used to treat autoimmune diseases such as rheumatoid arthritis.

6-Phosphogluconolactonase (EC 3.1.1.31, 6PGL, PGLS, systematic name 6-phospho-D-glucono-1,5-lactone lactonohydrolase) is a cytosolic enzyme found in all organisms that catalyzes the hydrolysis of 6-phosphogluconolactone to 6-phosphogluconic acid in the oxidative phase of the pentose phosphate pathway:

Glycerol-3-phosphate dehydrogenase (GPDH) is an enzyme that catalyzes the reversible redox conversion of dihydroxyacetone phosphate to sn-glycerol 3-phosphate.

Adenylosuccinate lyase is an enzyme that in humans is encoded by the ADSL gene.
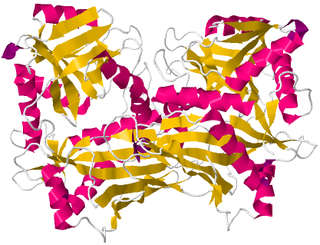
Branched-chain amino acid aminotransferase (BCAT), also known as branched-chain amino acid transaminase, is an aminotransferase enzyme (EC 2.6.1.42) which acts upon branched-chain amino acids (BCAAs). It is encoded by the BCAT2 gene in humans. The BCAT enzyme catalyzes the conversion of BCAAs and α-ketoglutarate into branched chain α-keto acids and glutamate.

Electron-transferring-flavoprotein dehydrogenase is an enzyme that transfers electrons from electron-transferring flavoprotein in the mitochondrial matrix, to the ubiquinone pool in the inner mitochondrial membrane. It is part of the electron transport chain. The enzyme is found in both prokaryotes and eukaryotes and contains a flavin and FE-S cluster. In humans, it is encoded by the ETFDH gene. Deficiency in ETF dehydrogenase causes the human genetic disease multiple acyl-CoA dehydrogenase deficiency.
In enzymology, a 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase (EC 1.3.1.28) is an enzyme that catalyzes the chemical reaction
In enzymology, carbon monoxide dehydrogenase (CODH) (EC 1.2.7.4) is an enzyme that catalyzes the chemical reaction
In enzymology, a proline racemase is an enzyme that catalyzes the chemical reaction

The enzyme lactoylglutathione lyase (EC 4.4.1.5, also known as glyoxalase I) catalyzes the isomerization of hemithioacetal adducts, which are formed in a spontaneous reaction between a glutathionyl group and aldehydes such as methylglyoxal.
In molecular biology, the PyrD leader is a cis-regulatory RNA element found at the 5' of the PyrC mRNA in Pseudomonadota. The PyrD gene encodes dihydroorotate dehydrogenase, an enzyme involved in pyrimidine biosynthesis. The PyrD leader regulates expression of PyrD. Translation initiation can occur at more than one different site within this leader sequence, under high cytidine triphosphate or guanosine triphosphate conditions the translation initiation site is upstream of that used under low CTP/GTP conditions, additional cytosine residues are incorporated into the mRNA resulting in the formation of an RNA hairpin. This hairpin blocks ribosome-binding at the Shine-Dalgarno sequence, and therefore blocks expression of PyrD. Under low CTP/GTP conditions the initiation site is further downstream and does not result in hairpin formation, so the ribosome can bind to the Shine-Dalgarno sequence and PyrD is expressed.
Soluble quinoprotein glucose dehydrogenase is an enzyme with systematic name D-glucose:acceptor oxidoreductase. This enzyme catalyses the following chemical reaction

Class 2 dihydroorotate dehydrogenases is an enzyme with systematic name (S)-dihydroorotate:quinone oxidoreductase. This enzyme catalyses the electron transfer from dihydroorotate to a quinone :
Lactocepin is an enzyme. This enzyme catalyses the following chemical reaction

Oxidosqualene cyclases (OSC) are enzymes involved in cyclization reactions of 2,3-oxidosqualene to form sterols or triterpenes.













