
A bacteriophage, also known informally as a phage, is a virus that infects and replicates within bacteria and archaea. The term was derived from "bacteria" and the Greek φαγεῖν, meaning "to devour". Bacteriophages are composed of proteins that encapsulate a DNA or RNA genome, and may have structures that are either simple or elaborate. Their genomes may encode as few as four genes and as many as hundreds of genes. Phages replicate within the bacterium following the injection of their genome into its cytoplasm.

Bacterial conjugation is the transfer of genetic material between bacterial cells by direct cell-to-cell contact or by a bridge-like connection between two cells. This takes place through a pilus. It is a parasexual mode of reproduction in bacteria.

Enterobacteria phage λ is a bacterial virus, or bacteriophage, that infects the bacterial species Escherichia coli. It was discovered by Esther Lederberg in 1950. The wild type of this virus has a temperate life cycle that allows it to either reside within the genome of its host through lysogeny or enter into a lytic phase, during which it kills and lyses the cell to produce offspring. Lambda strains, mutated at specific sites, are unable to lysogenize cells; instead, they grow and enter the lytic cycle after superinfecting an already lysogenized cell.

Escherichia coli ( ESH-ə-RIK-ee-ə KOH-lye) is a gram-negative, facultative anaerobic, rod-shaped, coliform bacterium of the genus Escherichia that is commonly found in the lower intestine of warm-blooded organisms. Most E. coli strains are harmless, but some serotypes such as EPEC, and ETEC are pathogenic and can cause serious food poisoning in their hosts, and are occasionally responsible for food contamination incidents that prompt product recalls. Most strains are part of the normal microbiota of the gut and are harmless or even beneficial to humans (although these strains tend to be less studied than the pathogenic ones). For example, some strains of E. coli benefit their hosts by producing vitamin K2 or by preventing the colonization of the intestine by pathogenic bacteria. These mutually beneficial relationships between E. coli and humans are a type of mutualistic biological relationship — where both the humans and the E. coli are benefitting each other. E. coli is expelled into the environment within fecal matter. The bacterium grows massively in fresh fecal matter under aerobic conditions for three days, but its numbers decline slowly afterwards.

In molecular biology and genetics, transformation is the genetic alteration of a cell resulting from the direct uptake and incorporation of exogenous genetic material from its surroundings through the cell membrane(s). For transformation to take place, the recipient bacterium must be in a state of competence, which might occur in nature as a time-limited response to environmental conditions such as starvation and cell density, and may also be induced in a laboratory.

Transduction is the process by which foreign DNA is introduced into a cell by a virus or viral vector. An example is the viral transfer of DNA from one bacterium to another and hence an example of horizontal gene transfer. Transduction does not require physical contact between the cell donating the DNA and the cell receiving the DNA, and it is DNase resistant. Transduction is a common tool used by molecular biologists to stably introduce a foreign gene into a host cell's genome.

Escherichia virus T4 is a species of bacteriophages that infect Escherichia coli bacteria. It is a double-stranded DNA virus in the subfamily Tevenvirinae of the family Straboviridae. T4 is capable of undergoing only a lytic life cycle and not the lysogenic life cycle. The species was formerly named T-even bacteriophage, a name which also encompasses, among other strains, Enterobacteria phage T2, Enterobacteria phage T4 and Enterobacteria phage T6.
DNA gyrase, or simply gyrase, is an enzyme within the class of topoisomerase and is a subclass of Type II topoisomerases that reduces topological strain in an ATP dependent manner while double-stranded DNA is being unwound by elongating RNA-polymerase or by helicase in front of the progressing replication fork. It is the only known enzyme to actively contribute negative supercoiling to DNA, while it also is capable of relaxing positive supercoils. It does so by looping the template to form a crossing, then cutting one of the double helices and passing the other through it before releasing the break, changing the linking number by two in each enzymatic step. This process occurs in bacteria, whose single circular DNA is cut by DNA gyrase and the two ends are then twisted around each other to form supercoils. Gyrase is also found in eukaryotic plastids: it has been found in the apicoplast of the malarial parasite Plasmodium falciparum and in chloroplasts of several plants. Bacterial DNA gyrase is the target of many antibiotics, including nalidixic acid, novobiocin, albicidin, and ciprofloxacin.

The phi X 174 bacteriophage is a single-stranded DNA (ssDNA) virus that infects Escherichia coli. This virus was isolated in 1935 by Nicolas Bulgakov in Félix d'Hérelle's laboratory at the Pasteur Institute, from samples collected in Paris sewers. Its characterization and the study of its replication mechanism were carried out from the 1950s onwards. It was the first DNA-based genome to be sequenced. This work was completed by Fred Sanger and his team in 1977. In 1962, Walter Fiers and Robert Sinsheimer had already demonstrated the physical, covalently closed circularity of ΦX174 DNA. Nobel prize winner Arthur Kornberg used ΦX174 as a model to first prove that DNA synthesized in a test tube by purified enzymes could produce all the features of a natural virus, ushering in the age of synthetic biology. In 1972–1974, Jerard Hurwitz, Sue Wickner, and Reed Wickner with collaborators identified the genes required to produce the enzymes to catalyze conversion of the single stranded form of the virus to the double stranded replicative form. In 2003, it was reported by Craig Venter's group that the genome of ΦX174 was the first to be completely assembled in vitro from synthesized oligonucleotides. The ΦX174 virus particle has also been successfully assembled in vitro. In 2012, it was shown how its highly overlapping genome can be fully decompressed and still remain functional.

Bacteriophage T7 is a bacteriophage, a virus that infects bacteria. It infects most strains of Escherichia coli and relies on these hosts to propagate. Bacteriophage T7 has a lytic life cycle, meaning that it destroys the cell it infects. It also possesses several properties that make it an ideal phage for experimentation: its purification and concentration have produced consistent values in chemical analyses; it can be rendered noninfectious by exposure to UV light; and it can be used in phage display to clone RNA binding proteins.
Recombineering is a genetic and molecular biology technique based on homologous recombination systems, as opposed to the older/more common method of using restriction enzymes and ligases to combine DNA sequences in a specified order. Recombineering is widely used for bacterial genetics, in the generation of target vectors for making a conditional mouse knockout, and for modifying DNA of any source often contained on a bacterial artificial chromosome (BAC), among other applications.
Allan McCulloch Campbell was an American microbiologist and geneticist and the Barbara Kimball Browning Professor Emeritus in the Department of Biology at Stanford University. His pioneering work on Lambda phage helped to advance molecular biology in the late 20th century. An important collaborator and member of his laboratory at Stanford University was biochemist Alice del Campillo Campbell, his wife.

Esther Miriam Zimmer Lederberg was an American microbiologist and a pioneer of bacterial genetics. She discovered the bacterial virus lambda phage and the bacterial fertility factor F, devised the first implementation of replica plating, and furthered the understanding of the transfer of genes between bacteria by specialized transduction.
P1 is a temperate bacteriophage that infects Escherichia coli and some other bacteria. When undergoing a lysogenic cycle the phage genome exists as a plasmid in the bacterium unlike other phages that integrate into the host DNA. P1 has an icosahedral head containing the DNA attached to a contractile tail with six tail fibers. The P1 phage has gained research interest because it can be used to transfer DNA from one bacterial cell to another in a process known as transduction. As it replicates during its lytic cycle it captures fragments of the host chromosome. If the resulting viral particles are used to infect a different host the captured DNA fragments can be integrated into the new host's genome. This method of in vivo genetic engineering was widely used for many years and is still used today, though to a lesser extent. P1 can also be used to create the P1-derived artificial chromosome cloning vector which can carry relatively large fragments of DNA. P1 encodes a site-specific recombinase, Cre, that is widely used to carry out cell-specific or time-specific DNA recombination by flanking the target DNA with loxP sites.
A P1-derived artificial chromosome, or PAC, is a DNA construct derived from the DNA of P1 bacteriophages and Bacterial artificial chromosome. It can carry large amounts of other sequences for a variety of bioengineering purposes in bacteria. It is one type of the efficient cloning vector used to clone DNA fragments in Escherichia coli cells.

A toxin-antitoxin system consists of a "toxin" and a corresponding "antitoxin", usually encoded by closely linked genes. The toxin is usually a protein while the antitoxin can be a protein or an RNA. Toxin-antitoxin systems are widely distributed in prokaryotes, and organisms often have them in multiple copies. When these systems are contained on plasmids – transferable genetic elements – they ensure that only the daughter cells that inherit the plasmid survive after cell division. If the plasmid is absent in a daughter cell, the unstable antitoxin is degraded and the stable toxic protein kills the new cell; this is known as 'post-segregational killing' (PSK).
The CTXφ bacteriophage is a filamentous bacteriophage. It is a positive-strand DNA virus with single-stranded DNA (ssDNA).

Daisy Roulland-Dussoix was a Swiss molecular microbiologist. She was one of the discoverers of restriction enzymes during her doctoral studies. There is controversy over whether she should have received the 1978 Nobel prize in Physiology and Medicine, which was awarded to Hamilton O. Smith, Daniel Nathans, and Werner Arber.
Escherichia virus CC31, formerly known as Enterobacter virus CC31, is a dsDNA bacteriophage of the subfamily Tevenvirinae responsible for infecting the bacteria family of Enterobacteriaceae. It is one of two discovered viruses of the genus Karamvirus, diverging away from the previously discovered T4virus, as a clonal complex (CC). CC31 was first isolated from Escherichia coli B strain S/6/4 and is primarily associated with Escherichia, even though is named after Enterobacter.
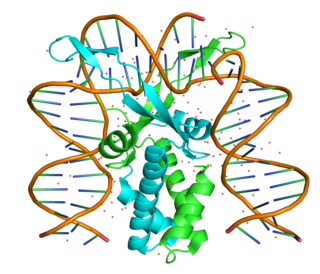
The integration host factor (IHF) is a bacterial DNA binding protein complex that facilitates genetic recombination, replication, and transcription by binding to specific DNA sequences and bending the DNA. It also facilitates the integration of foreign DNA into the host genome. It is a heterodimeric complex composed of two homologous subunits IHFalpha and IHFbeta.













