
Oxidative phosphorylation or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis.

Selenocysteine is the 21st proteinogenic amino acid. Selenoproteins contain selenocysteine residues. Selenocysteine is an analogue of the more common cysteine with selenium in place of the sulfur.

Malate dehydrogenase (EC 1.1.1.37) (MDH) is an enzyme that reversibly catalyzes the oxidation of malate to oxaloacetate using the reduction of NAD+ to NADH. This reaction is part of many metabolic pathways, including the citric acid cycle. Other malate dehydrogenases, which have other EC numbers and catalyze other reactions oxidizing malate, have qualified names like malate dehydrogenase (NADP+).

In biochemistry, mixed acid fermentation is the metabolic process by which a six-carbon sugar is converted into a complex and variable mixture of acids. It is an anaerobic (non-oxygen-requiring) fermentation reaction that is common in bacteria. It is characteristic for members of the Enterobacteriaceae, a large family of Gram-negative bacteria that includes E. coli.
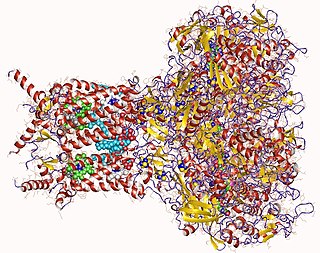
Formate dehydrogenases are a set of enzymes that catalyse the oxidation of formate to carbon dioxide, donating the electrons to a second substrate, such as NAD+ in formate:NAD+ oxidoreductase (EC 1.17.1.9) or to a cytochrome in formate:ferricytochrome-b1 oxidoreductase (EC 1.2.2.1). This family of enzymes has attracted attention as inspiration or guidance on methods for the carbon dioxide fixation, relevant to global warming.
In enzymology, a malate oxidase (EC 1.1.3.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a quinoprotein glucose dehydrogenase is an enzyme that catalyzes the chemical reaction
In enzymology, an aminobutyraldehyde dehydrogenase (EC 1.2.1.19) is an enzyme that catalyzes the chemical reaction
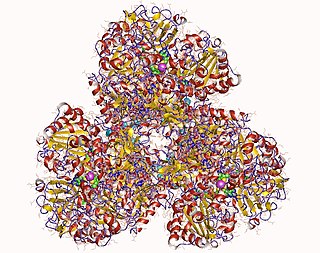
In enzymology, a formate dehydrogenase (cytochrome) (EC 1.2.2.1) is an enzyme that catalyzes the chemical reaction

In enzymology, a nicotinate dehydrogenase is an enzyme that catalyzes the chemical reaction

Cytochrome c nitrite reductase (ccNiR) is a bacterial enzyme that catalyzes the six electron reduction of nitrite to ammonia; an important step in the biological nitrogen cycle. The enzyme catalyses the second step in the two step conversion of nitrate to ammonia, which allows certain bacteria to use nitrite as a terminal electron acceptor, rather than oxygen, during anaerobic conditions. During this process, ccNiR draws electrons from the quinol pool, which are ultimately provided by a dehydrogenase such as formate dehydrogenase or hydrogenase. These dehydrogenases are responsible for generating a proton motive force.
The Arc system is a two-component system found in some bacteria that regulates gene expression in faculatative anaerobes such as Escheria coli. Two-component system means that it has a sensor molecule and a response regulator. Arc is an abbreviation for Anoxic Redox Control system. Arc systems are instrumental in maintaining energy metabolism during transcription of bacteria. The ArcA response regulator looks at growth conditions and expresses genes to best suit the bacteria. The Arc B sensor kinase, which is a tripartite protein, is membrane bound and can autophosphorylate.
The fnr gene of Escherichia coli encodes a transcriptional activator (FNR) which is required for the expression of a number of genes involved in anaerobic respiratory pathways. The FNR protein of E. coli is an oxygen – responsive transcriptional regulator required for the switch from aerobic to anaerobic metabolism.
"Type III mutants, originally frdB, were designated fnr because they were defective in fumarate and nitrate reduction and impaired in their ability to produce gas." - Lambden and Guest, 1976 Journal of General Microbiology97, 145-160

Methionine sulfoxide is the organic compound with the formula CH3S(O)CH2CH2CH(NH2)CO2H. It is an amino acid that occurs naturally although it is formed post-translationally.
The Formate-Nitrite Transporter (FNT) Family belongs to the Major Intrinsic Protein (MIP) Superfamily. FNT family members have been sequenced from Gram-negative and Gram-positive bacteria, archaea, yeast, plants and lower eukaryotes. The prokaryotic proteins of the FNT family probably function in the transport of the structurally related compounds, formate and nitrite.

Although it is toxic in large doses, selenium is an essential micronutrient for animals. In plants, it sometimes occurs in toxic amounts as forage, e.g. locoweed. Selenium is a component of the amino acids selenocysteine and selenomethionine. In humans, selenium is a trace element nutrient that functions as cofactor for glutathione peroxidases and certain forms of thioredoxin reductase. Selenium-containing proteins are produced from inorganic selenium via the intermediacy of selenophosphate (PSeO33−).
Quinate/shikimate dehydrogenase (EC 1.1.1.282, YdiB) is an enzyme with systematic name L-quinate:NAD(P)+ 3-oxidoreductase. This enzyme catalyses the following chemical reaction
Formate dehydrogenase-N (EC 1.1.5.6, Fdh-N, FdnGHI, nitrate-inducible formate dehydrogenase, formate dehydrogenase N, FDH-N, nitrate inducible Fdn, nitrate inducible formate dehydrogenase) is an enzyme with systematic name formate:quinone oxidoreductase. This enzyme catalyses the following chemical reaction

Fumarate reductase (quinol) (EC 1.3.5.4, QFR,FRD, menaquinol-fumarate oxidoreductase, quinol:fumarate reductase) is an enzyme with systematic name succinate:quinone oxidoreductase. This enzyme catalyzes the following chemical reaction:
O-phosphoseryl-tRNASec kinase is an enzyme with systematic name ATP:L-seryl-tRNASec O-phosphotransferase. This enzyme catalyses the following chemical reaction











