
Haplogroup J-M304, also known as J, is a human Y-chromosome DNA haplogroup. It is believed to have evolved in Western Asia. The clade spread from there during the Neolithic, primarily into North Africa, the Horn of Africa, Socotra, the Caucasus, Europe, West Asia, Central Asia, South Asia, and Southeast Asia.
Haplogroup E-M96 is a human Y-chromosome DNA haplogroup. It is one of the two main branches of the older and ancestral haplogroup DE, the other main branch being haplogroup D. The E-M96 clade is divided into two main subclades: the more common E-P147, and the less common E-M75.

Haplogroup F, also known as F-M89 and previously as Haplogroup FT is a very common Y-chromosome haplogroup. The clade and its subclades constitute over 90% of paternal lineages outside of Africa.
Haplogroup K or K-M9 is a human Y-chromosome DNA haplogroup. A sublineage of haplogroup IJK, K-M9, and its descendant clades represent a geographically widespread and diverse haplogroup. The lineages have long been found among males on every continent except Antarctica.

Haplogroup N (M231) is a Y-chromosome DNA haplogroup defined by the presence of the single-nucleotide polymorphism (SNP) marker M231.
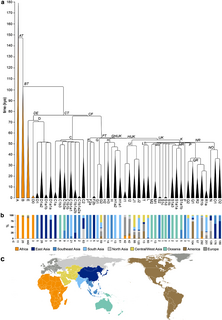
In human genetics, a human Y-chromosome DNA haplogroup is a haplogroup defined by mutations in the non-recombining portions of DNA from the male-specific Y chromosome. Many people within a haplogroup share similar numbers of short tandem repeats (STRs) and types of mutations called single-nucleotide polymorphisms (SNPs).
In human genetics, Haplogroup O-M268, also known as O1b, is a Y-chromosome DNA haplogroup. Haplogroup O-M268 is a primary subclade of haplogroup O-F265, itself a primary descendant branch of Haplogroup O-M175.
In human genetics, Haplogroup O-M119 is a Y-chromosome DNA haplogroup. Haplogroup O-M119 is a descendant branch of haplogroup O-F265, one of two extant primary subclades of Haplogroup O-M175. The same clade previously has been labeled as O-MSY2.2.

Haplogroup O-M176 or O1b2 is a human Y-chromosome DNA haplogroup. It is best known for its part in the settlement of Korea and Japan. It is a descendant of Haplogroup O-P31, and it has been estimated to share a most recent common ancestor with its nearest outgroup, Haplogroup O-K18, approximately 28,300 [95% CI 26,100 <-> 30,500] years before present, approximately 29,100 years before present, or approximately 31,108 years before present.
Haplogroup I-M253, also known as I1, is a Y chromosome haplogroup. The genetic markers confirmed as identifying I-M253 are the SNPs M253,M307.2/P203.2, M450/S109, P30, P40, L64, L75, L80, L81, L118, L121/S62, L123, L124/S64, L125/S65, L157.1, L186, and L187. It is a primary branch of Haplogroup I-M170 (I*).
Haplogroup I-M438, also known as I2, is a human DNA Y-chromosome haplogroup, a subclade of Haplogroup I-M170. Haplogroup I-M438 originated some time around 26,000–31,000 BCE. It originated in Europe and developed into several main subgroups : I2-M438*, I2a-L460, I2b-L415 and I2c-L596. The haplogroup can be found all over Europe and reaches its maximum frequency in the Dinaric Alps (Balkans) via founder effect. Examples of basal I-M438* have been found in males from Crete and Sicily.
Haplogroup DE is a human Y-chromosome DNA haplogroup. It is defined by the single nucleotide polymorphism (SNP) mutations, or UEPs, M1(YAP), M145(P205), M203, P144, P153, P165, P167, P183. DE is unique because it is distributed in several geographically distinct clusters. An immediate subclade, haplogroup D, is mainly found in East Asia, parts of Central Asia, and the Andaman Islands, but also sporadically in West Africa and West Asia. The other immediate subclade, haplogroup E, is common in Africa, and to a lesser extent the Middle East and Europe.

Haplogroup CT is a human Y chromosome haplogroup, defining one of the major paternal lineages of humanity.
Haplogroup K2, also known as K-M526 and formerly known as K(xLT) and MNOPS, is a human Y-DNA haplogroup.
In human population genetics, haplogroups define the major lineages of direct paternal (male) lines back to a shared common ancestor in Africa.
HaplogroupGHIJK, defined by the SNPs M3658, F1329, PF2622, and YSC0001299, is a common Y-chromosome haplogroup. This macrohaplogroup and its subclades contain the vast majority of the world's existing male population.
Haplogroup P1, also known as P-M45 and K2b2a, is a Y-chromosome DNA haplogroup in human genetics. Defined by the SNPs M45 and PF5962, P1 is a primary branch (subclade) of P.

Haplogroup K2a is a human Y-chromosome DNA haplogroup. K2a is a primary subclade of haplogroup K2 (M526), which in turn is a primary descendant of haplogroup K (M9). Its sole primary descendant is haplogroup K-M2313.
As with all modern European nations, a large degree of 'biological continuity' exists between Bosnians and Bosniaks and their ancient predecessors with Y chromosomal lineages testifying to predominantly Paleolithic European ancestry. Studies based on bi-allelic markers of the NRY have shown the three main ethnic groups of Bosnia and Herzegovina to share, in spite of some quantitative differences, a large fraction of the same ancient gene pool distinct for the region. Analysis of autosomal STRs have moreover revealed no significant difference between the population of Bosnia and Herzegovina and neighbouring populations.

Haplogroup D or D-CTS3946 is a Y-chromosome haplogroup. Like its relative distant sibling, haplogroup E-M96, D-CTS3946 has the YAP+ unique-event polymorphism, which defines their parent, haplogroup DE.









