Influenza A virus (IAV) is a pathogen that causes the flu in birds and some mammals, including humans. It is an RNA virus whose subtypes have been isolated from wild birds. Occasionally, it is transmitted from wild to domestic birds, and this may cause severe disease, outbreaks, or human influenza pandemics.

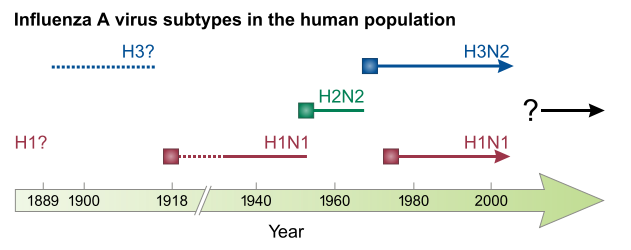
Antigenic shift is the process by which two or more different strains of a virus, or strains of two or more different viruses, combine to form a new subtype having a mixture of the surface antigens of the two or more original strains. The term is often applied specifically to influenza, as that is the best-known example, but the process is also known to occur with other viruses, such as visna virus in sheep. Antigenic shift is a specific case of reassortment or viral shift that confers a phenotypic change.

Influenza hemagglutinin (HA) or haemagglutinin[p] is a homotrimeric glycoprotein found on the surface of influenza viruses and is integral to its infectivity.

Orthomyxoviridae is a family of negative-sense RNA viruses. It includes seven genera: Alphainfluenzavirus, Betainfluenzavirus, Gammainfluenzavirus, Deltainfluenzavirus, Isavirus, Thogotovirus, and Quaranjavirus. The first four genera contain viruses that cause influenza in birds and mammals, including humans. Isaviruses infect salmon; the thogotoviruses are arboviruses, infecting vertebrates and invertebrates. The Quaranjaviruses are also arboviruses, infecting vertebrates (birds) and invertebrates (arthropods).

Influenza A virus subtype H5N1 (A/H5N1) is a subtype of the influenza A virus which can cause illness in humans and many other species. A bird-adapted strain of H5N1, called HPAI A(H5N1) for highly pathogenic avian influenza virus of type A of subtype H5N1, is the highly pathogenic causative agent of H5N1 flu, commonly known as avian influenza. It is enzootic in many bird populations, especially in Southeast Asia. One strain of HPAI A(H5N1) is spreading globally after first appearing in Asia. It is epizootic and panzootic, killing tens of millions of birds and spurring the culling of hundreds of millions of others to stem its spread. Many references to "bird flu" and H5N1 in the popular media refer to this strain.
The Hong Kong flu, also known as the 1968 flu pandemic, was a flu pandemic whose outbreak in 1968 and 1969 killed between one and four million people globally. It is among the deadliest pandemics in history, and was caused by an H3N2 strain of the influenza A virus. The virus was descended from H2N2 through antigenic shift, a genetic process in which genes from multiple subtypes are reassorted to form a new virus.

In virology, influenza A virus subtype H1N1 (A/H1N1) is a subtype of influenza A virus. Major outbreaks of H1N1 strains in humans include the 1918 Spanish flu pandemic, the 1977 Russian flu pandemic and the 2009 swine flu pandemic. It is an orthomyxovirus that contains the glycoproteins hemagglutinin (H) and neuraminidase (N), antigens whose subtypes are used to classify the strains of the virus as H1N1, H1N2 etc. Hemagglutinin causes red blood cells to clump together and binds the virus to the infected cell. Neuraminidase is a type of glycoside hydrolase enzyme which helps to move the virus particles through the infected cell and assist in budding from the host cells.

An influenza pandemic is an epidemic of an influenza virus that spreads across a large region and infects a large proportion of the population. There have been six major influenza epidemics in the last 140 years, with the 1918 flu pandemic being the most severe; this is estimated to have been responsible for the deaths of 50–100 million people. The most recent, the 2009 swine flu pandemic, resulted in under 300,000 deaths and is considered relatively mild. These pandemics occur irregularly.

Influenza A virus subtype H3N2 (A/H3N2) is a subtype of viruses that causes influenza (flu). H3N2 viruses can infect birds and mammals. In birds, humans, and pigs, the virus has mutated into many strains. In years in which H3N2 is the predominant strain, there are more hospitalizations.

Transmission and infection of H5N1 from infected avian sources to humans has been a concern since the first documented case of human infection in 1997, due to the global spread of H5N1 that constitutes a pandemic threat.

Influenza B virus is the only species in the genus Betainfluenzavirus in the virus family Orthomyxoviridae.
H3N8 is a subtype of the species Influenza A virus that is endemic in birds, horses and dogs. It is the main cause of equine influenza and is also known as equine influenza virus. In 2011, it was reported to have been found in seals. Cats have been experimentally infected with the virus, leading to clinical signs, shedding of the virus and infection of other cats. In 2022 and 2023, three people in China were infected with H3N8, with one fatality, marking the first time a human has died from this strain of flu.

H5N1 genetic structure is the molecular structure of the H5N1 virus's RNA.

H5N1 clinical trials are clinical trials concerning H5N1 vaccines, which are intended to provide immunization to influenza A virus subtype H5N1. They are intended to discover pharmacological effects and identify any adverse reactions the vaccines may achieve in humans.

Fujian flu refers to flu caused by either a Fujian human flu strain of the H3N2 subtype of the Influenza A virus or a Fujian bird flu strain of the H5N1 subtype of the Influenza A virus. These strains are named after Fujian, a coastal province in Southeast China.

The Goose Guangdong virus refers to the strain A/Goose/Guangdong/1/96 (Gs/Gd)-like H5N1 HPAI viruses. It is a strain of the Influenzavirus A subtype H5N1 virus that was first detected in a goose in Guangdong in 1996. It is an HPAI virus, meaning that it can kill a very high percentage of chickens in a flock in mere days. It is believed to be the immediate precursor of the current dominant strain of HPAI A(H5N1) that evolved from 1999 to 2002 creating the Z genotype that is spreading globally and is epizootic and panzootic, killing tens of millions of birds and spurring the culling of hundreds of millions of others to stem its spread.

Human mortality from H5N1 or the human fatality ratio from H5N1 or the case-fatality rate of H5N1 is the ratio of the number of confirmed human deaths resulting from confirmed cases of transmission and infection of H5N1 to the number of those confirmed cases. For example, if there are 100 confirmed cases of humans infected with H5N1 and 10 die, then there is a 10% human fatality ratio. H5N1 flu is a concern due to the global spread of H5N1 that constitutes a pandemic threat. The majority of H5N1 flu cases have been reported in southeast and east Asia. The case-fatality rate is central to pandemic planning. Estimates of case-fatality (CF) rates for past influenza pandemics have ranged from to 2-3% for the 1918 pandemic to about 0.6% for the 1957 pandemic to 0.2% for the 1968 pandemic. As of 2008, the official World Health Organization estimate for the case-fatality rate for the outbreak of H5N1 avian influenza was approximately 60%. Public health officials in Ontario, Canada argue that the true case-fatality rate could be lower, pointing to studies suggesting it could be 14-33%, and warned that it was unlikely to be as low as the 0.1–0.4% rate that was built into many pandemic plans.

Influenza, commonly known as "the flu", is an infectious disease caused by influenza viruses. Symptoms range from mild to severe and often include fever, runny nose, sore throat, muscle pain, headache, coughing, and fatigue. These symptoms begin from one to four days after exposure to the virus and last for about 2–8 days. Diarrhea and vomiting can occur, particularly in children. Influenza may progress to pneumonia, which can be caused by the virus or by a subsequent bacterial infection. Other complications of infection include acute respiratory distress syndrome, meningitis, encephalitis, and worsening of pre-existing health problems such as asthma and cardiovascular disease.
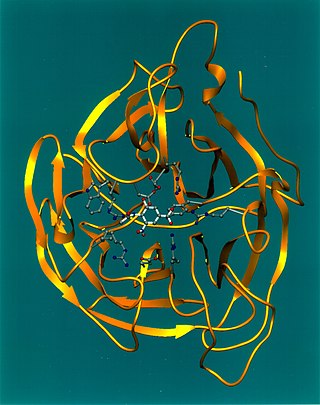
Viral neuraminidase is a type of neuraminidase found on the surface of influenza viruses that enables the virus to be released from the host cell. Neuraminidases are enzymes that cleave sialic acid groups from glycoproteins. Neuraminidase inhibitors are antiviral agents that inhibit influenza viral neuraminidase activity and are of major importance in the control of influenza.

A H5N1 vaccine is an influenza vaccine intended to provide immunization to influenza A virus subtype H5N1.