Related Research Articles
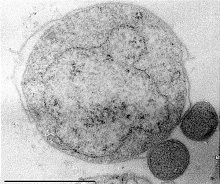
Nanoarchaeum equitans is a species of marine archaea that was discovered in 2002 in a hydrothermal vent off the coast of Iceland on the Kolbeinsey Ridge by Karl Stetter. It has been proposed as the first species in a new phylum, and is the only species within the genus Nanoarchaeum. Strains of this microbe were also found on the Sub-polar Mid Oceanic Ridge, and in the Obsidian Pool in Yellowstone National Park. Since it grows in temperatures approaching boiling, at about 80 degrees Celsius, it is considered to be a thermophile. It grows best in environments with a pH of 6, and a salinity concentration of 2%. Nanoarchaeum appears to be an obligate symbiont on the archaeon Ignicoccus; it must be in contact with the host organism to survive. Nanoarchaeum equitans cannot synthesize lipids but obtains them from its host. Its cells are only 400 nm in diameter, making it the smallest known living organism, and the smallest known archaeon.
A hyperthermophile is an organism that thrives in extremely hot environments—from 60 °C (140 °F) upwards. An optimal temperature for the existence of hyperthermophiles is often above 80 °C (176 °F). Hyperthermophiles are often within the domain Archaea, although some bacteria are also able to tolerate extreme temperatures. Some of these bacteria are able to live at temperatures greater than 100 °C, deep in the ocean where high pressures increase the boiling point of water. Many hyperthermophiles are also able to withstand other environmental extremes, such as high acidity or high radiation levels. Hyperthermophiles are a subset of extremophiles. Their existence may support the possibility of extraterrestrial life, showing that life can thrive in environmental extremes.
Methanogens are microorganisms that produce methane as a metabolic byproduct in hypoxic conditions. They are prokaryotic and belong to the domain Archaea. All known methanogens are members of the archaeal phylum Euryarchaeota. Methanogens are common in wetlands, where they are responsible for marsh gas, and in the digestive tracts of animals such as ruminants and many humans, where they are responsible for the methane content of belching in ruminants and flatulence in humans. In marine sediments, the biological production of methane, also termed methanogenesis, is generally confined to where sulfates are depleted, below the top layers. Moreover, methanogenic archaea populations play an indispensable role in anaerobic wastewater treatments. Others are extremophiles, found in environments such as hot springs and submarine hydrothermal vents as well as in the "solid" rock of Earth's crust, kilometers below the surface.
Archaeoglobus is a genus of the phylum Euryarchaeota. Archaeoglobus can be found in high-temperature oil fields where they may contribute to oil field souring.
Methanopyrus is a genus of the Methanopyraceae.

Sulfolobus is a genus of microorganism in the family Sulfolobaceae. It belongs to the archaea domain.

A thermoacidophile is an extremophilic microorganism that is both thermophilic and acidophilic; i.e., it can grow under conditions of high temperature and low pH. The large majority of thermoacidophiles are archaea or bacteria, though occasional eukaryotic examples have been reported. Thermoacidophiles can be found in hot springs and solfataric environments, within deep sea vents, or in other environments of geothermal activity. They also occur in polluted environments, such as in acid mine drainage.
Methanosarcina acetivorans is a versatile methane producing microbe which is found in such diverse environments as oil wells, trash dumps, deep-sea hydrothermal vents, and oxygen-depleted sediments beneath kelp beds. Only M. acetivorans and microbes in the genus Methanosarcina use all three known metabolic pathways for methanogenesis. Methanosarcinides, including M. acetivorans, are also the only archaea capable of forming multicellular colonies, and even show cellular differentiation. The genome of M. acetivorans is one of the largest archaeal genomes ever sequenced. Furthermore, one strain of M. acetivorans, M. a. C2A, has been identified to possess an F-type ATPase along with an A-type ATPase.

Methanosarcina is a genus of euryarchaeote archaea that produce methane. These single-celled organisms are known as anaerobic methanogens that produce methane using all three metabolic pathways for methanogenesis. They live in diverse environments where they can remain safe from the effects of oxygen, whether on the earth's surface, in groundwater, in deep sea vents, and in animal digestive tracts. Methanosarcina grow in colonies.

Nitrosopumilus maritimus is an extremely common archaeon living in seawater. It is the first member of the Group 1a Nitrososphaerota to be isolated in pure culture. Gene sequences suggest that the Group 1a Nitrososphaerota are ubiquitous with the oligotrophic surface ocean and can be found in most non-coastal marine waters around the planet. It is one of the smallest living organisms at 0.2 micrometers in diameter. Cells in the species N. maritimus are shaped like peanuts and can be found both as individuals and in loose aggregates. They oxidize ammonia to nitrite and members of N. maritimus can oxidize ammonia at levels as low as 10 nanomolar, near the limit to sustain its life. Archaea in the species N. maritimus live in oxygen-depleted habitats. Oxygen needed for ammonia oxidation might be produced by novel pathway which generates oxygen and dinitrogen. N. maritimus is thus among organisms which are able to produce oxygen in dark.
Methanobrevibacter smithii is the predominant archaeon in the microbiota of the human gut. M. smithii has a coccobacillus shape. It plays an important role in the efficient digestion of polysaccharides by consuming the end products of bacterial fermentation. Methanobrevibacter smithii is a single-celled microorganism from the Archaea domain. M. smithii is a methanogen, and a hydrogenotroph that recycles the hydrogen by combining it with carbon dioxide to methane. The removal of hydrogen by M. smithii is thought to allow an increase in the extraction of energy from nutrients by shifting bacterial fermentation to more oxidized end products.

Archaea is a domain of single-celled organisms. These microorganisms lack cell nuclei and are therefore prokaryotes. Archaea were initially classified as bacteria, receiving the name archaebacteria, but this term has fallen out of use.
Thermococcus celer is a Gram-negative, spherical-shaped archaeon of the genus Thermococcus. The discovery of T. celer played an important role in rerooting the tree of life when T. celer was found to be more closely related to methanogenic Archaea than to other phenotypically similar thermophilic species. T. celer was the first archaeon discovered to house a circularized genome. Several type strains of T. celer have been identified: Vu13, ATCC 35543, and DSM 2476.
Methanocaldococcus jannaschii is a thermophilic methanogenic archaean in the class Methanococci. It was the first archaeon, and third organism, to have its complete genome sequenced. The sequencing identified many genes unique to the archaea. Many of the synthesis pathways for methanogenic cofactors were worked out biochemically in this organism, as were several other archaeal-specific metabolic pathways.
Methanogenium frigidum is a psychrophilic, H2-using methanogen from Ace Lake, Antarctica.
Thermoplasma volcanium is a moderate thermoacidophilic archaea isolated from acidic hydrothermal vents and solfatara fields. It contains no cell wall and is motile. It is a facultative anaerobic chemoorganoheterotroph. No previous phylogenetic classifications have been made for this organism. Thermoplasma volcanium reproduces asexually via binary fission and is nonpathogenic.
Picrophilus torridus is a species of Archaea described in 1996. Picrophilus torridus was found in soil near a hot spring in Hokkaido, Japan. The pH of the soil was less than 0.5. P. torridus also has one of the smallest genomes found among organisms that are free-living and are non-parasitic and a high coding density, meaning that the majority of its genes are coding regions and provide instructions for building proteins. The current research suggests the two hostile conditions favored by P. torridus have exerted selective pressure towards having a small and compact genome, which is less likely to be damaged by the harsh environment.

Methanohalophilus mahii is an obligately anaerobic, methylotrophic, methanogenic cocci-shaped archaeon of the genus Methanohalophilus that can be found in high salinity aquatic environments. The name Methanohalophilus is said to be derived from methanum meaning "methane" in Latin; halo meaning "salt" in Greek; and mahii meaning "of Mah" in Latin, after R.A. Mah, who did substantial amounts of research on aerobic and methanogenic microbes. The proper word in ancient Greek for "salt" is however hals (ἅλς). The specific strain type was designated SLP and is currently the only identified strain of this species.
Methanocaldococcussp. FS406-22 is an archaea in the genus Methanocaldococcus. It is an anaerobic, piezophilic, diazotrophic, hyperthermophilic marine archaeon. This strain is notable for fixing nitrogen at the highest known temperature of nitrogen fixers recorded to date. The 16S rRNA gene of Methanocaldococcus sp. FS406-22, is almost 100% similar to that of Methanocaldococcus jannaschii, a non-nitrogen fixer.
"Candidatus Thorarchaeota", or simply Thorarchaeota, is a phylum within the superphylum Asgard archaea. The Asgard superphylum represents the closest prokaryotic relatives of eukaryotes. Since there is such a close relation between the two different domains, it provides further evidence to the two-domain tree of life theory which states that eukaryotes branched from the archaeal domain. Asgard archaea are single cell marine microbes that contain branch like appendages and have genes that are similar to eukarya. The asgard archaea superphylum is composed of Thorarchaeota, Lokiarchaeota, Odinarchaeota, and Heimdallarchaeota. Thorarchaeota were first identified from the sulfate-methane transition zone in tidewater sediments. Thorarcheota are widely distributed in marine and freshwater sediments.
References
- ↑ Franzmann, P.D.; Springer, N.; Ludwig, W.; Conway De Macario, E.; Rohde, M. (1992). "A Methanogenic Archaeon from Ace Lake, Antarctica: Methanococcoides burtonii sp. nov". Systematic and Applied Microbiology. 15 (4): 573–581. doi:10.1016/S0723-2020(11)80117-7. ISSN 0723-2020.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Franzmann, P.D.; Springer, N.; Ludwig, W.; Conway de Macario, E.; et al. (1992). "A Methanogenic Archeon from Ace Lake, Antarctica: Methanococcoides burtonii sp. nov". Syst. Appl. Microbiol. 15 (4): 573–581. doi:10.1016/s0723-2020(11)80117-7.
- 1 2 3 4 5 6 7 8 9 Goodchild, Amber; Saunders, N.; Ertan, H.; Raftery, M.; Guilhaus, M.; Curmi, P.; Cavicchioli, R. (2004). "A proteomic determination of cold adaptation in the Antarctic archeon, Methanococcoides burtonii". Molecular Microbiology. 53 (1): 309–321. doi: 10.1111/j.1365-2958.2004.04130.x . PMID 15225324. S2CID 22384609.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Nichols, David; Miller, M.; Davies, N.; Goodchild, A.; et al. (2004). "Cold Adaptation in the Antarctic Archaeon Methanococcoides burtonii Involves Membrane Lipid Unsaturation". Journal of Bacteriology. 186 (24): 8508–8515. doi:10.1128/JB.186.24.8508-8515.2004. PMC 532414 . PMID 15576801.
- 1 2 3 Saunders, Neil; Thomas, T.; Curmi, P.M.G.; Mattick, J.S.; et al. (2003). "Mechanisms of Thermal Adaptation Revealed From the Genomes of the Antarctic Archaea Methanogenium frigidum and Methanococcoides burtonii". Genome Research. 13 (7): 1580–1588. doi:10.1101/gr.1180903. PMC 403754 . PMID 12805271.
- ↑ Goodchild, Amber; Cavicchioli, Ricardo; Gilhaus, Michael; Raftery, Mark; et al. (2005). "Cold Adaptation of the Antarctic Archaeon, Methanococcoides burtonii Assessed by Proteomics Using ICAT". Journal of Proteome Research. 4 (2): 473–480. doi:10.1021/pr049760p. PMID 15822924.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 Allen, Michelle; Lauro, F.; Wiliams, T.; Burg, D.; et al. (2009). "The genome sequence of the psychrophilic arcaeon, Methanococcoides burtonii: the role of genome evolution in cold adaption". ISME Journal. 3 (9): 1012–1035. doi: 10.1038/ismej.2009.45 . PMID 19404327.