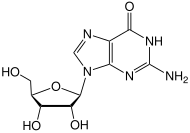
The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG sites occur with high frequency in genomic regions called CpG islands.

A glioma is a type of primary tumor that starts in the glial cells of the brain or spinal cord. They are cancerous but some are extremely slow to develop. Gliomas comprise about 30 percent of all brain tumors and central nervous system tumours, and 80 percent of all malignant brain tumours.

Glioblastoma, previously known as glioblastoma multiforme (GBM), is the most aggressive and most common type of cancer that originates in the brain, and has a very poor prognosis for survival. Initial signs and symptoms of glioblastoma are nonspecific. They may include headaches, personality changes, nausea, and symptoms similar to those of a stroke. Symptoms often worsen rapidly and may progress to unconsciousness.

A neoplasm is a type of abnormal and excessive growth of tissue. The process that occurs to form or produce a neoplasm is called neoplasia. The growth of a neoplasm is uncoordinated with that of the normal surrounding tissue, and persists in growing abnormally, even if the original trigger is removed. This abnormal growth usually forms a mass, which may be called a tumour or tumor.
Malignant transformation is the process by which cells acquire the properties of cancer. This may occur as a primary process in normal tissue, or secondarily as malignant degeneration of a previously existing benign tumor.

DNA mismatch repair (MMR) is a system for recognizing and repairing erroneous insertion, deletion, and mis-incorporation of bases that can arise during DNA replication and recombination, as well as repairing some forms of DNA damage.
DNA mismatch repair protein Msh2 also known as MutS homolog 2 or MSH2 is a protein that in humans is encoded by the MSH2 gene, which is located on chromosome 2. MSH2 is a tumor suppressor gene and more specifically a caretaker gene that codes for a DNA mismatch repair (MMR) protein, MSH2, which forms a heterodimer with MSH6 to make the human MutSα mismatch repair complex. It also dimerizes with MSH3 to form the MutSβ DNA repair complex. MSH2 is involved in many different forms of DNA repair, including transcription-coupled repair, homologous recombination, and base excision repair.

DNA mismatch repair protein Mlh1 or MutL protein homolog 1 is a protein that in humans is encoded by the MLH1 gene located on chromosome 3. The gene is commonly associated with hereditary nonpolyposis colorectal cancer. Orthologs of human MLH1 have also been studied in other organisms including mouse and the budding yeast Saccharomyces cerevisiae.

Cilengitide is a molecule designed and synthesized at the Technical University Munich in collaboration with Merck KGaA in Darmstadt. It is based on the cyclic peptide cyclo(-RGDfV-), which is selective for αv integrins, which are important in angiogenesis, and other aspects of tumor biology. Hence, it is under investigation for the treatment of glioblastoma, where it may act by inhibiting angiogenesis, and influencing tumor invasion and proliferation.

6-O-Methylguanine is a derivative of the nucleobase guanine in which a methyl group is attached to the oxygen atom. It base-pairs to thymine rather than cytosine, causing a G:C to A:T transition in DNA.

Temozolomide, sold under the brand name Temodar among others, is an anticancer medication used to treat brain tumors such as glioblastoma and anaplastic astrocytoma. It is taken by mouth or via intravenous infusion.
Genome instability refers to a high frequency of mutations within the genome of a cellular lineage. These mutations can include changes in nucleic acid sequences, chromosomal rearrangements or aneuploidy. Genome instability does occur in bacteria. In multicellular organisms genome instability is central to carcinogenesis, and in humans it is also a factor in some neurodegenerative diseases such as amyotrophic lateral sclerosis or the neuromuscular disease myotonic dystrophy.

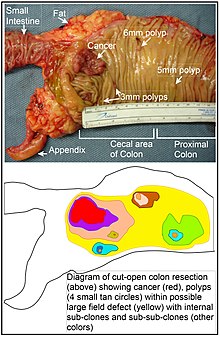
Field cancerization or field effect is a biological process in which large areas of cells at a tissue surface or within an organ are affected by carcinogenic alterations. The process arises from exposure to an injurious environment, often over a lengthy period.

Cancer epigenetics is the study of epigenetic modifications to the DNA of cancer cells that do not involve a change in the nucleotide sequence, but instead involve a change in the way the genetic code is expressed. Epigenetic mechanisms are necessary to maintain normal sequences of tissue specific gene expression and are crucial for normal development. They may be just as important, if not even more important, than genetic mutations in a cell's transformation to cancer. The disturbance of epigenetic processes in cancers, can lead to a loss of expression of genes that occurs about 10 times more frequently by transcription silencing than by mutations. As Vogelstein et al. points out, in a colorectal cancer there are usually about 3 to 6 driver mutations and 33 to 66 hitchhiker or passenger mutations. However, in colon tumors compared to adjacent normal-appearing colonic mucosa, there are about 600 to 800 heavily methylated CpG islands in the promoters of genes in the tumors while these CpG islands are not methylated in the adjacent mucosa. Manipulation of epigenetic alterations holds great promise for cancer prevention, detection, and therapy. In different types of cancer, a variety of epigenetic mechanisms can be perturbed, such as the silencing of tumor suppressor genes and activation of oncogenes by altered CpG island methylation patterns, histone modifications, and dysregulation of DNA binding proteins. There are several medications which have epigenetic impact, that are now used in a number of these diseases.
Antineoplastic resistance, often used interchangeably with chemotherapy resistance, is the resistance of neoplastic (cancerous) cells, or the ability of cancer cells to survive and grow despite anti-cancer therapies. In some cases, cancers can evolve resistance to multiple drugs, called multiple drug resistance.
Neuroepigenetics is the study of how epigenetic changes to genes affect the nervous system. These changes may effect underlying conditions such as addiction, cognition, and neurological development.
Generally, in progression to cancer, hundreds of genes are silenced or activated. Although silencing of some genes in cancers occurs by mutation, a large proportion of carcinogenic gene silencing is a result of altered DNA methylation. DNA methylation causing silencing in cancer typically occurs at multiple CpG sites in the CpG islands that are present in the promoters of protein coding genes.
DNA methylation in cancer plays a variety of roles, helping to change the healthy cells by regulation of gene expression to a cancer cells or a diseased cells disease pattern. One of the most widely studied DNA methylation dysregulation is the promoter hypermethylation where the CPGs islands in the promoter regions are methylated contributing or causing genes to be silenced.
CpG island hypermethylation is a phenomenon that is important for the regulation of gene expression in cancer cells, as an epigenetic control aberration responsible for gene inactivation. Hypermethylation of CpG islands has been described in almost every type of tumor.
Pharmacoepigenetics is an emerging field that studies the underlying epigenetic marking patterns that lead to variation in an individual's response to medical treatment.