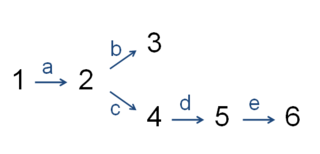Peptidoglycan or murein is a unique large macromolecule, a polysaccharide, consisting of sugars and amino acids that forms a mesh-like peptidoglycan layer outside the plasma membrane, the rigid cell wall characteristic of most bacteria. The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is an oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer. Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer which is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply, a technique carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.

Teichoic acids are bacterial copolymers of glycerol phosphate or ribitol phosphate and carbohydrates linked via phosphodiester bonds.

N-Acetylmuramic acid is an organic compound with the chemical formula C
11H
19NO
8. It is a monomer of peptidoglycan in most bacterial cell walls, which is built from alternating units of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid, cross-linked by oligopeptides at the lactic acid residue of MurNAc.

N-Acetylglucosamine (GlcNAc) is an amide derivative of the monosaccharide glucose. It is a secondary amide between glucosamine and acetic acid. It is significant in several biological systems.

In biochemistry, mixed acid fermentation is the metabolic process by which a six-carbon sugar is converted into a complex and variable mixture of acids. It is an anaerobic (non-oxygen-requiring) fermentation reaction that is common in bacteria. It is characteristic for members of the Enterobacteriaceae, a large family of Gram-negative bacteria that includes E. coli.

The transsulfuration pathway is a metabolic pathway involving the interconversion of cysteine and homocysteine through the intermediate cystathionine. Two transsulfurylation pathways are known: the forward and the reverse.

In enzymology, an UDP-N-acetylmuramate dehydrogenase (EC 1.3.1.98) is an enzyme that catalyzes the chemical reaction
In enzymology, an UDP-N-acetylglucosamine 2-epimerase is an enzyme that catalyzes the chemical reaction

Isocitrate lyase, or ICL, is an enzyme in the glyoxylate cycle that catalyzes the cleavage of isocitrate to succinate and glyoxylate. Together with malate synthase, it bypasses the two decarboxylation steps of the tricarboxylic acid cycle and is used by bacteria, fungi, and plants.

In enzymology, N-acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25), also known as GlcNAc-6-phosphate deacetylase or NagA, is an enzyme that catalyzes the deacetylation of N-acetylglucosamine-6-phosphate (GlcNAc-6-P) to glucosamine-6-phosphate (GlcN-6-P):

In enzymology, glucosamine-phosphate N-acetyltransferase (GNA) is an enzyme that catalyzes the transfer of an acetyl group from acetyl-CoA to the primary amine in glucosamide-6-phosphate, generating a free CoA and N-acetyl-D-glucosamine-6-phosphate.

In enzymology, a malate synthase (EC 2.3.3.9) is an enzyme that catalyzes the chemical reaction

In enzymology, an UDP-N-acetylglucosamine 1-carboxyvinyltransferase is an enzyme that catalyzes the first committed step in peptidoglycan biosynthesis of bacteria:
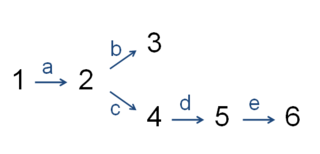
In enzymology, the committed step is an effectively irreversible enzymatic reaction that occurs at a branch point during the biosynthesis of some molecules. As the name implies, after this step, the molecules are "committed" to the pathway and will ultimately end up in the pathway's final product. The first committed step should not be confused with the rate-determining step, which is the slowest step in a reaction or pathway. However, it is sometimes the case that the first committed step is in fact the rate-determining step as well.
The Arc system is a two-component system found in some bacteria that regulates gene expression in faculatative anaerobes such as Escheria coli. Two-component system means that it has a sensor molecule and a response regulator. Arc is an abbreviation for Anoxic Redox Control system. Arc systems are instrumental in maintaining energy metabolism during transcription of bacteria. The ArcA response regulator looks at growth conditions and expresses genes to best suit the bacteria. The Arc B sensor kinase, which is a tripartite protein, is membrane bound and can autophosphorylate.
The bacterial cell wall provides strength and rigidity to counteract internal osmotic pressure, and protection against the environment. The peptidoglycan layer gives the cell wall its strength, and helps maintain the overall shape of the cell. The basic peptidoglycan structure of both Gram-positive and Gram-negative bacteria comprises a sheet of glycan chains connected by short cross-linking polypeptides. Biosynthesis of peptidoglycan is a multi-step process comprising three main stages:
- formation of UDP-N-acetylmuramic acid (UDPMurNAc) from N-acetylglucosamine (GlcNAc).
- addition of a short polypeptide chain to the UDPMurNAc.
- addition of a second GlcNAc to the disaccharide-pentapeptide building block and transport of this unit through the cytoplasmic membrane and incorporation into the growing peptidoglycan layer.
N,N'-diacetylchitobiose phosphorylase is an enzyme with the systematic name N,N'-diacetylchitobiose:phosphate N-acetyl-D-glucosaminyltransferase. This enzyme was found in the genus Vibrio initially but has now been found to be taken up by Escherichia coli as well as many other bacteria. One study shows that Escherichia coli can replicate on a medium that is just composed of GlcNAc a product of phosphorylation of N,N'-diacetylchitobiose as the sole source of carbon. Because E. coli can go on this medium, the enzyme is present. The enzyme has also been found in multiple eukaryotic cells as well, especially in eukaryotes that make chitin and break chitin down. It is believed that N,N'-diacetylchitobiose phosphorylase is an integral part of the phosphoenolpyruvate:glucose phosphotransferase system (PTS). It is assumed that it is involved with Enzyme Complex II of the PTS and is involved with the synthesis of chitin. The enzyme is specific for N,N'-diacetylchitobiose.
Anhydro-N-acetylmuramic acid kinase (EC 2.7.1.170, anhMurNAc kinase, AnmK) is an enzyme with systematic name ATP:1,6-anhydro-N-acetyl-beta-muramate 6-phosphotransferase. This enzyme catalyses the following chemical reaction
UDP-N-acetylglucosamine kinase is an enzyme with systematic name ATP:UDP-N-acetyl-alpha-D-glucosamine 3'-phosphotransferase. This enzyme catalyses the following chemical reaction
UDP-N-acetylglucosamine—undecaprenyl-phosphate N-acetylglucosaminephosphotransferase is an enzyme with systematic name UDP-N-acetyl-alpha-D-glucosamine:ditrans,octacis-undecaprenyl phosphate N-acetyl-alpha-D-glucosaminephosphotransferase. This enzyme catalyses the following chemical reaction