| sn-glycerol-1-phosphate dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 1.1.1.261 | ||||||||
| CAS no. | 204594-18-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
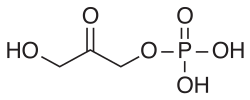
In enzymology, sn-glycerol-1-phosphate dehydrogenase (EC 1.1.1.261) is an enzyme that catalyzes the chemical reaction
The two substrates of this enzyme are (S)-glyceryl 1-phosphate and oxidised nicotinamide adenine dinucleotide (NAD+). Its products are glycerone phosphate, reduced NADH, and a proton. The enzyme can also use the alternative cofactor, nicotinamide adenine dinucleotide phosphate. [1] [2]
This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is sn-glycerol-1-phosphate:NAD(P)+ 2-oxidoreductase. This enzyme is also called glycerol-1-phosphate dehydrogenase [NAD(P)+].
G-1-P dehydrogenase is responsible for the formation of sn-glycerol 1-phosphate, the backbone of the membrane phospholipids of Archaea. The gene encoding glycerol-1-phosphate dehydrogenase has been detected in all the archaeal species and has not been found in any bacterial or eukaryal species. sn-glycerol 1-phosphate produced by this enzyme is the most fundamental difference by which Archaea and bacteria are discriminated.
The enzyme sn-glycerol-1-phosphate dehydrogenase, usually having 394 amino acids, was also identified in bacteria. More than 5700 sequences have been published in GenBank (September 2023) in a different bacteria, including such well-known ones as Bacillus subtilis (GenBank: AOR99168.1).