
Structural biology, as defined by the Journal of Structural Biology, deals with structural analysis of living material at every level of organization.
The Protein Data Bank (PDB) is a database for the three-dimensional structural data of large biological molecules such as proteins and nucleic acids, which is overseen by the Worldwide Protein Data Bank (wwPDB). These structural data are obtained and deposited by biologists and biochemists worldwide through the use of experimental methodologies such as X-ray crystallography, NMR spectroscopy, and, increasingly, cryo-electron microscopy. All submitted data are reviewed by expert biocurators and, once approved, are made freely available on the Internet under the CC0 Public Domain Dedication. Global access to the data is provided by the websites of the wwPDB member organisations.

Structural bioinformatics is the branch of bioinformatics that is related to the analysis and prediction of the three-dimensional structure of biological macromolecules such as proteins, RNA, and DNA. It deals with generalizations about macromolecular 3D structures such as comparisons of overall folds and local motifs, principles of molecular folding, evolution, binding interactions, and structure/function relationships, working both from experimentally solved structures and from computational models. The term structural has the same meaning as in structural biology, and structural bioinformatics can be seen as a part of computational structural biology. The main objective of structural bioinformatics is the creation of new methods of analysing and manipulating biological macromolecular data in order to solve problems in biology and generate new knowledge.
BioJava is an open-source software project dedicated to provide Java tools to process biological data. BioJava is a set of library functions written in the programming language Java for manipulating sequences, protein structures, file parsers, Common Object Request Broker Architecture (CORBA) interoperability, Distributed Annotation System (DAS), access to AceDB, dynamic programming, and simple statistical routines. BioJava supports a range of data, starting from DNA and protein sequences to the level of 3D protein structures. The BioJava libraries are useful for automating many daily and mundane bioinformatics tasks such as to parsing a Protein Data Bank (PDB) file, interacting with Jmol and many more. This application programming interface (API) provides various file parsers, data models and algorithms to facilitate working with the standard data formats and enables rapid application development and analysis.
A chemical file format is a type of data file which is used specifically for depicting molecular data. One of the most widely used is the chemical table file format, which is similar to Structure Data Format (SDF) files. They are text files that represent multiple chemical structure records and associated data fields. The XYZ file format is a simple format that usually gives the number of atoms in the first line, a comment on the second, followed by a number of lines with atomic symbols and cartesian coordinates. The Protein Data Bank Format is commonly used for proteins but is also used for other types of molecules. There are many other types which are detailed below. Various software systems are available to convert from one format to another.
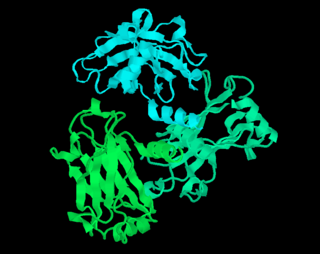
RasMol is a computer program written for molecular graphics visualization intended and used mainly to depict and explore biological macromolecule structures, such as those found in the Protein Data Bank (PDB).
The European Bioinformatics Institute (EMBL-EBI) is an intergovernmental organization (IGO) which, as part of the European Molecular Biology Laboratory (EMBL) family, focuses on research and services in bioinformatics. It is located on the Wellcome Genome Campus in Hinxton near Cambridge, and employs over 600 full-time equivalent (FTE) staff.
The Protein Data Bank (PDB) file format is a textual file format describing the three-dimensional structures of molecules held in the Protein Data Bank, now succeeded by the mmCIF format. The PDB format accordingly provides for description and annotation of protein and nucleic acid structures including atomic coordinates, secondary structure assignments, as well as atomic connectivity. In addition experimental metadata are stored. The PDB format is the legacy file format for the Protein Data Bank which has kept data on biological macromolecules in the newer PDBx/mmCIF file format since 2014.
The EM Data Bank or Electron Microscopy Data Bank (EMDB) collects 3D EM maps and associated experimental data determined using electron microscopy of biological specimens. It was established in 2002 at the MSD/PDBe group of the European Bioinformatics Institute (EBI), where the European site of the EMDataBank.org consortium is located. As of 2015, the resource contained over 2,600 entries with a mean resolution of 15Å.

Helen Miriam Berman is a Board of Governors Professor of Chemistry and Chemical Biology at Rutgers University and a former director of the RCSB Protein Data Bank. A structural biologist, her work includes structural analysis of protein-nucleic acid complexes, and the role of water in molecular interactions. She is also the founder and director of the Nucleic Acid Database, and led the Protein Structure Initiative Structural Genomics Knowledgebase.
In biology, a protein structure database is a database that is modeled around the various experimentally determined protein structures. The aim of most protein structure databases is to organize and annotate the protein structures, providing the biological community access to the experimental data in a useful way. Data included in protein structure databases often includes three-dimensional coordinates as well as experimental information, such as unit cell dimensions and angles for x-ray crystallography determined structures. Though most instances, in this case either proteins or a specific structure determinations of a protein, also contain sequence information and some databases even provide means for performing sequence based queries, the primary attribute of a structure database is structural information, whereas sequence databases focus on sequence information, and contain no structural information for the majority of entries. Protein structure databases are critical for many efforts in computational biology such as structure based drug design, both in developing the computational methods used and in providing a large experimental dataset used by some methods to provide insights about the function of a protein.
Biological data visualization is a branch of bioinformatics concerned with the application of computer graphics, scientific visualization, and information visualization to different areas of the life sciences. This includes visualization of sequences, genomes, alignments, phylogenies, macromolecular structures, systems biology, microscopy, and magnetic resonance imaging data. Software tools used for visualizing biological data range from simple, standalone programs to complex, integrated systems.

David S. Goodsell, is an associate professor at the Scripps Research Institute and research professor at Rutgers University, New Jersey. He is especially known for his watercolor paintings of cell interiors.
The Re-referenced Protein Chemical shift Database (RefDB) is an NMR spectroscopy database of carefully corrected or re-referenced chemical shifts, derived from the BioMagResBank (BMRB). The database was assembled by using a structure-based chemical shift calculation program to calculate expected protein (1)H, (13)C and (15)N chemical shifts from X-ray or NMR coordinate data of previously assigned proteins reported in the BMRB. The comparison is automatically performed by a program called SHIFTCOR. The RefDB database currently provides reference-corrected chemical shift data on more than 2000 assigned peptides and proteins. Data from the database indicates that nearly 25% of BMRB entries with (13)C protein assignments and 27% of BMRB entries with (15)N protein assignments require significant chemical shift reference readjustments. Additionally, nearly 40% of protein entries deposited in the BioMagResBank appear to have at least one assignment error. Users may download, search or browse the database through a number of methods available through the RefDB website. RefDB provides a standard chemical shift resource for biomolecular NMR spectroscopists, wishing to derive or compute chemical shift trends in peptides and proteins.

WeNMR is a worldwide e-Infrastructure for NMR spectroscopy and structural biology. It is the largest virtual Organization in the life sciences and is supported by EGI.

Macromolecular structure validation is the process of evaluating reliability for 3-dimensional atomic models of large biological molecules such as proteins and nucleic acids. These models, which provide 3D coordinates for each atom in the molecule, come from structural biology experiments such as x-ray crystallography or nuclear magnetic resonance (NMR). The validation has three aspects: 1) checking on the validity of the thousands to millions of measurements in the experiment; 2) checking how consistent the atomic model is with those experimental data; and 3) checking consistency of the model with known physical and chemical properties.
Stephen Kevin Burley is a British-born scientist, naturalized in both Canada and the United States, specializing in oncology and structural biology. He is a University Professor and Henry Rutgers Chair at Rutgers, The State University of New Jersey. Burley directs the RCSB Protein Data Bank, the Center for Integrative Proteomics Research, and the Institute for Quantitative Biomedicine.
The Biological Magnetic Resonance Data Bank is an open access repository of nuclear magnetic resonance (NMR) spectroscopic data from peptides, proteins, nucleic acids and other biologically relevant molecules. The database is operated by the University of Wisconsin–Madison and is supported by the National Library of Medicine. The BMRB is part of the Research Collaboratory for Structural Bioinformatics and, since 2006, it is a partner in the Worldwide Protein Data Bank (wwPDB). The repository accepts NMR spectral data from laboratories around the world and, once the data is validated, it is available online at the BMRB website. The database has also an ftp site, where data can be downloaded in the bulk. The BMRB has two mirror sites, one at the Protein Database Japan (PDBj) at Osaka University and one at the Magnetic Resonance Research Center (CERM) at the University of Florence in Italy. The site at Japan accepts and processes data depositions.
Protein Data Bank in Europe – Knowledge Base (PDBe-KB) is a community-driven, open-access, integrated resource whose mission is to place macromolecular structure data in their biological context and to make them accessible to the scientific community in order to support fundamental and translational research and education. It is part of the European Bioinformatics Institute (EMBL-EBI), based at the Wellcome Genome Campus, Hinxton, Cambridgeshire, England.

Gaetano T. Montelione is an American biophysical chemist, Professor of Chemistry and Chemical Biology, and Constellation Endowed Chair in Structural Bioinformatics at Rensselaer Polytechnic Institute in Troy, NY.








