| (S)-mandelate dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 1.1.99.31 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
In enzymology, (S)-mandelate dehydrogenase (EC 1.1.99.31) (MDH), is an enzyme that catalyzes the chemical reaction.
 + acceptor + acceptor | = | 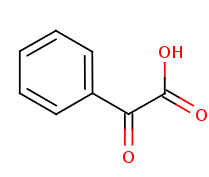 + reduced acceptor + reduced acceptor |
|---|---|---|
| (S)-Mandelate | 2-oxo-2-phenylacetate | |
| (S)-2-hydroxy-2-phenylacetate + acceptor ⇌ 2-oxo-2-phenylacetate + reduced acceptor | ||
Thus, the two substrates of this enzyme are (S)-2-hydroxy-2-phenylacetate and acceptor, whereas its two products are 2-oxo-2-phenylacetate and reduced acceptor.
This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-OH group of donor with other acceptors. The systematic name of this enzyme class is (S)-2-hydroxy-2-phenylacetate:acceptor 2-oxidoreductase.
This enzyme transfers the electron pair from FMNH2 to a component of the electron transport chain, most probably ubiquinone [1,2]. It is part of a metabolic pathway in Pseudomonads that allows these organisms to utilize mandelic acid, derivatized from the common soil metabolite amygdalin, as the sole source of carbon and energy. The enzyme has a large active-site pocket and preferentially binds substrates with longer sidechains, e.g. 2-hydroxyoctanoate rather than 2-hydroxybutyrate. It also prefers substrates that, like (S)-mandelate, have beta unsaturation, e.g. (indol-3-yl)glycolate compared with (indol-3-yl)lactate. Esters of mandelate, such as methyl (S)-mandelate, are also substrates. [1]