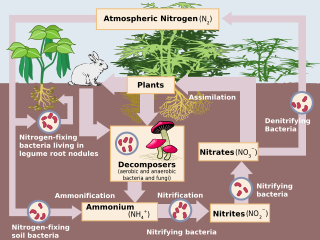
Nitrification is the biological oxidation of ammonia to nitrite followed by the oxidation of the nitrite to nitrate occurring through separate organisms or direct ammonia oxidation to nitrate in comammox bacteria. The transformation of ammonia to nitrite is usually the rate limiting step of nitrification. Nitrification is an important step in the nitrogen cycle in soil. Nitrification is an aerobic process performed by small groups of autotrophic bacteria and archaea.

Nitrosomonas is a genus of Gram-negative bacteria, belonging to the Betaproteobacteria. It is one of the five genera of ammonia-oxidizing bacteria and, as an obligate chemolithoautotroph, uses ammonia as an energy source and carbon dioxide as a carbon source in presence of oxygen. Nitrosomonas are important in the global biogeochemical nitrogen cycle, since they increase the bioavailability of nitrogen to plants and in the denitrification, which is important for the release of nitrous oxide, a powerful greenhouse gas. This microbe is photophobic, and usually generate a biofilm matrix, or form clumps with other microbes, to avoid light. Nitrosomonas can be divided into six lineages: the first one includes the species Nitrosomonas europea, Nitrosomonas eutropha, Nitrosomonas halophila, and Nitrosomonas mobilis. The second lineage presents the species Nitrosomonas communis, N. sp. I and N. sp. II, meanwhile the third lineage includes only Nitrosomonas nitrosa. The fourth lineage includes the species Nitrosomonas ureae and Nitrosomonas oligotropha and the fifth and sixth lineages include the species Nitrosomonas marina, N. sp. III, Nitrosomonas estuarii and Nitrosomonas cryotolerans.
Nitrifying bacteria are chemolithotrophic organisms that include species of genera such as Nitrosomonas, Nitrosococcus, Nitrobacter, Nitrospina, Nitrospira and Nitrococcus. These bacteria get their energy from the oxidation of inorganic nitrogen compounds. Types include ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB). Many species of nitrifying bacteria have complex internal membrane systems that are the location for key enzymes in nitrification: ammonia monooxygenase, hydroxylamine oxidoreductase, and nitrite oxidoreductase.
Nitrospira translate into “a nitrate spiral” is a genus of bacteria within the monophyletic clade of the Nitrospirota phylum. The first member of this genus was described 1986 by Watson et al. isolated from the Gulf of Maine. The bacterium was named Nitrospira marina. Populations were initially thought to be limited to marine ecosystems, but it was later discovered to be well-suited for numerous habitats, including activated sludge of wastewater treatment systems, natural biological marine settings, water circulation biofilters in aquarium tanks, terrestrial systems, fresh and salt water ecosystems, and hot springs. Nitrospira is a ubiquitous bacterium that plays a role in the nitrogen cycle by performing nitrite oxidation in the second step of nitrification. Nitrospira live in a wide array of environments including but not limited to, drinking water systems, waste treatment plants, rice paddies, forest soils, geothermal springs, and sponge tissue. Despite being abundant in many natural and engineered ecosystems Nitrospira are difficult to culture, so most knowledge of them is from molecular and genomic data. However, due to their difficulty to be cultivated in laboratory settings, the entire genome was only sequenced in one species, Nitrospira defluvii. In addition, Nitrospira bacteria's 16s rRNA sequences are too dissimilar to use for PCR primers, thus some members go unnoticed. In addition, members of Nitrospira with the capabilities to perform complete nitrification has also been discovered and cultivated.
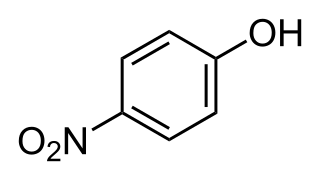
4-Nitrophenol is a phenolic compound that has a nitro group at the opposite position of the hydroxyl group on the benzene ring.

Squalene monooxygenase is a eukaryotic enzyme that uses NADPH and diatomic oxygen to oxidize squalene to 2,3-oxidosqualene. Squalene epoxidase catalyzes the first oxygenation step in sterol biosynthesis and is thought to be one of the rate-limiting enzymes in this pathway. In humans, squalene epoxidase is encoded by the SQLE gene. Several eukaryote genomes lack a squalene monooxygenase encoding gene, but instead encode an alternative squalene epoxidase that performs the same task.
In enzymology, a 2-nitrophenol 2-monooxygenase (EC 1.14.13.31) is an enzyme that catalyzes the chemical reaction
In enzymology, a 4-nitrophenol 2-monooxygenase (EC 1.14.13.29) is an enzyme that catalyzes the chemical reaction

Dopamine beta-hydroxylase (DBH), also known as dopamine beta-monooxygenase, is an enzyme that in humans is encoded by the DBH gene. Dopamine beta-hydroxylase catalyzes the conversion of dopamine to norepinephrine.
In enzymology, a trans-cinnamate 4-monooxygenase (EC 1.14.14.91) is an enzyme that catalyzes the chemical reaction
In enzymology, a p-benzoquinone reductase (NADPH) (EC 1.6.5.6) is an enzyme that catalyzes the chemical reaction

Dimethylaniline monooxygenase [N-oxide-forming] 1 is an enzyme that in humans is encoded by the FMO1 gene.

Dimethylaniline monooxygenase [N-oxide-forming] 5 is an enzyme that in humans is encoded by the FMO5 gene.

Cytochrome P450 2A13 is a protein that in humans is encoded by the CYP2A13 gene.
2-Hydroxy-1,4-benzoxazin-3-one monooxygenase (EC 1.14.13.140, BX5 (gene), CYP71C3 (gene)) is an enzyme with systematic name 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one,NAD(P)H:oxygen oxidoreductase (N-hydroxylating). This enzyme catalyses the following chemical reaction
6-hydroxy-3-succinoylpyridine 3-monooxygenase (EC 1.14.13.163, 6-hydroxy-3-succinoylpyridine hydroxylase, hspA (gene), hspB (gene)) is an enzyme with systematic name 4-(6-hydroxypyridin-3-yl)-4-oxobutanoate,NADH:oxygen oxidoreductase (3-hydroxylating, succinate semialdehyde releasing). This enzyme catalyses the following chemical reaction
4-nitrocatechol 4-monooxygenase (EC 1.14.13.166) is an enzyme with systematic name 4-nitrocatechol,NAD(P)H:oxygen 4-oxidoreductase (4-hydroxylating, nitrite-forming). This enzyme catalyses the following chemical reaction:
Ammonia monooxygenase (EC 1.14.99.39, AMO) is an enzyme, which catalyses the following chemical reaction

The flavin-containing monooxygenase (FMO) protein family specializes in the oxidation of xeno-substrates in order to facilitate the excretion of these compounds from living organisms. These enzymes can oxidize a wide array of heteroatoms, particularly soft nucleophiles, such as amines, sulfides, and phosphites. This reaction requires an oxygen, an NADPH cofactor, and an FAD prosthetic group. FMOs share several structural features, such as a NADPH binding domain, FAD binding domain, and a conserved arginine residue present in the active site. Recently, FMO enzymes have received a great deal of attention from the pharmaceutical industry both as a drug target for various diseases and as a means to metabolize pro-drug compounds into active pharmaceuticals. These monooxygenases are often misclassified because they share activity profiles similar to those of cytochrome P450 (CYP450), which is the major contributor to oxidative xenobiotic metabolism. However, a key difference between the two enzymes lies in how they proceed to oxidize their respective substrates; CYP enzymes make use of an oxygenated heme prosthetic group, while the FMO family utilizes FAD to oxidize its substrates.
Cytochrome P450, family 55, also known as CYP55, is a cytochrome P450 family in fungi supposed to derived from horizontal gene transfer of Actinomycetes CYP105 family member in the ancestor of all Dikarya. The first gene identified in this family is the CYP55A1 from Fusarium oxysporum encoding the NADPH dependent reductase of nitrous oxide.








