Histone methyltransferases (HMT) are histone-modifying enzymes, that catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. The attachment of methyl groups occurs predominantly at specific lysine or arginine residues on histones H3 and H4. Two major types of histone methyltranferases exist, lysine-specific and arginine-specific. In both types of histone methyltransferases, S-Adenosyl methionine (SAM) serves as a cofactor and methyl donor group.
The genomic DNA of eukaryotes associates with histones to form chromatin. The level of chromatin compaction depends heavily on histone methylation and other post-translational modifications of histones. Histone methylation is a principal epigenetic modification of chromatin that determines gene expression, genomic stability, stem cell maturation, cell lineage development, genetic imprinting, DNA methylation, and cell mitosis.
The androgen receptor (AR), also known as NR3C4, is a type of nuclear receptor that is activated by binding any of the androgenic hormones, including testosterone and dihydrotestosterone, in the cytoplasm and then translocating into the nucleus. The androgen receptor is most closely related to the progesterone receptor, and progestins in higher dosages can block the androgen receptor.
Histone methylation is a process by which methyl groups are transferred to amino acids of histone proteins that make up nucleosomes, which the DNA double helix wraps around to form chromosomes. Methylation of histones can either increase or decrease transcription of genes, depending on which amino acids in the histones are methylated, and how many methyl groups are attached. Methylation events that weaken chemical attractions between histone tails and DNA increase transcription because they enable the DNA to uncoil from nucleosomes so that transcription factor proteins and RNA polymerase can access the DNA. This process is critical for the regulation of gene expression that allows different cells to express different genes.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.

Proline-, glutamic acid- and leucine-rich protein 1 (PELP1) also known as modulator of non-genomic activity of estrogen receptor (MNAR) and transcription factor HMX3 is a protein that in humans is encoded by the PELP1 gene. is a transcriptional corepressor for nuclear receptors such as glucocorticoid receptors and a coactivator for estrogen receptors.
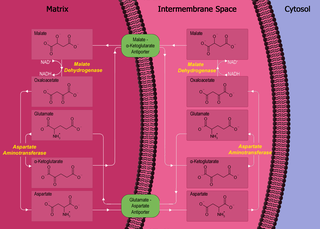
The malate-aspartate shuttle is a biochemical system for translocating electrons produced during glycolysis across the semipermeable inner membrane of the mitochondrion for oxidative phosphorylation in eukaryotes. These electrons enter the electron transport chain of the mitochondria via reduction equivalents to generate ATP. The shuttle system is required because the mitochondrial inner membrane is impermeable to NADH, the primary reducing equivalent of the electron transport chain. To circumvent this, malate carries the reducing equivalents across the membrane.
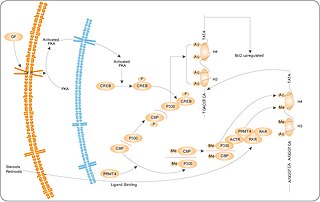
Protein arginine N-methyltransferase-4 (PRMT4/CARM1) methylation of arginine residues within proteins plays a critical key role in transcriptional regulation. PRMT4 binds to the classes of transcriptional activators known as p160 and CBP/p300. The modified forms of these proteins are involved in stimulation of gene expression via steroid hormone receptors. Significantly, PRMT4 methylates core histones H3 and H4, which are also targets of the histone acetylase activity of CBP/p300 coactivators. PRMT4 recruitment of chromatin by binding to coactivators increases histone methylation and enhances the accessibility of promoter regions for transcription. Methylation of the transcriptional coactivator CBP by PRMT4 inhibits binding to CREB and thereby partitions the limited cellular pool of CBP for steroid hormone receptor interaction.
The nuclear receptor coactivator 2 also known as NCoA-2 is a protein that in humans is encoded by the NCOA2 gene. NCoA-2 is also frequently called glucocorticoid receptor-interacting protein 1 (GRIP1), steroid receptor coactivator-2 (SRC-2), or transcriptional mediators/intermediary factor 2 (TIF2).

Enhancer of zeste homolog 2 (EZH2) is a histone-lysine N-methyltransferase enzyme encoded by EZH2 gene, that participates in histone methylation and, ultimately, transcriptional repression. EZH2 catalyzes the addition of methyl groups to histone H3 at lysine 27, by using the cofactor S-adenosyl-L-methionine. Methylation activity of EZH2 facilitates heterochromatin formation thereby silences gene function. Remodeling of chromosomal heterochromatin by EZH2 is also required during cell mitosis.
Protein arginine N-methyltransferase 1 is an enzyme that in humans is encoded by the PRMT1 gene. The HRMT1L2 gene encodes a protein arginine methyltransferase that functions as a histone methyltransferase specific for histone H4.
Four and a half LIM domains protein 2 also known as FHL-2 is a protein that in humans is encoded by the FHL2 gene. LIM proteins contain a highly conserved double zinc finger motif called the LIM domain.

Homeobox protein Nkx-3.1, also known as NKX3-1, NKX3, BAPX2, NKX3A and NKX3.1 is a protein that in humans is encoded by the NKX3-1 gene located on chromosome 8p. NKX3-1 is a prostatic tumor suppressor gene.

Protein UXT also known as androgen receptor trapped clone 27 (ART-27) protein is a protein that in humans is encoded by the UXT gene.

Methylosome protein 50 is a protein that in humans is encoded by the WDR77 gene.

E3 ubiquitin-protein ligase RNF14 is an enzyme that in humans is encoded by the RNF14 gene.

Melanoma-associated antigen 11 is a protein that in humans is encoded by the MAGEA11 gene. It is also involved in the androgen and progesterone receptor signaling pathways.

Malate dehydrogenase, cytoplasmic also known as malate dehydrogenase 1 is an enzyme that in humans is encoded by the MDH1 gene.
EPI-001 is the first inhibitor of the androgen receptor amino-terminal domain. The single stereoisomer of EPI-001, EPI-002, is a first-in-class drug that the USAN council assigned a new stem class "-aniten" and the generic name "ralaniten". This distinguishes the anitens novel molecular mechanism from anti androgens that bind the C-terminus ligand-binding domain and have the stem class "lutamide". EPI-001 and its stereoisomers and analogues were discovered by Marianne Sadar and Raymond Andersen, who co-founded the pharmaceutical company ESSA Pharma Inc for the clinical development of anitens for the treatment of castration-resistant prostate cancer (CRPC).
Protein methylation is a type of post-translational modification featuring the addition of methyl groups to proteins. It can occur on the nitrogen-containing side-chains of arginine and lysine, but also at the amino- and carboxy-termini of a number of different proteins. In biology, methyltransferases catalyze the methylation process, activated primarily by S-adenosylmethionine. Protein methylation has been most studied in histones, where the transfer of methyl groups from S-adenosyl methionine is catalyzed by histone methyltransferases. Histones that are methylated on certain residues can act epigenetically to repress or activate gene expression.
Minkui Luo is a biochemist and professor of biochemistry at Memorial Sloan Kettering Cancer Center. His research interests include chemical biology and the study of posttranslational modifications in epigenetic signaling, with an emphasis on protein methyltransferases.