
Nitric oxide synthases (NOSs) are a family of enzymes catalyzing the production of nitric oxide (NO) from L-arginine. NO is an important cellular signaling molecule. It helps modulate vascular tone, insulin secretion, airway tone, and peristalsis, and is involved in angiogenesis and neural development. It may function as a retrograde neurotransmitter. Nitric oxide is mediated in mammals by the calcium-calmodulin controlled isoenzymes eNOS and nNOS. The inducible isoform, iNOS, involved in immune response, binds calmodulin at physiologically relevant concentrations, and produces NO as an immune defense mechanism, as NO is a free radical with an unpaired electron. It is the proximate cause of septic shock and may function in autoimmune disease.

Clavulanic acid is a β-lactam drug that functions as a mechanism-based β-lactamase inhibitor. While not effective by itself as an antibiotic, when combined with penicillin-group antibiotics, it can overcome antibiotic resistance in bacteria that secrete β-lactamase, which otherwise inactivates most penicillins.

Methionine synthase also known as MS, MeSe, MTR is responsible for the regeneration of methionine from homocysteine. In humans it is encoded by the MTR gene (5-methyltetrahydrofolate-homocysteine methyltransferase). Methionine synthase forms part of the S-adenosylmethionine (SAMe) biosynthesis and regeneration cycle, and is the enzyme responsible for linking the cycle to one-carbon metabolism via the folate cycle. There are two primary forms of this enzyme, the Vitamin B12 (cobalamin)-dependent (MetH) and independent (MetE) forms, although minimal core methionine synthases that do not fit cleanly into either category have also been described in some anaerobic bacteria. The two dominant forms of the enzymes appear to be evolutionary independent and rely on considerably different chemical mechanisms. Mammals and other higher eukaryotes express only the cobalamin-dependent form. In contrast, the distribution of the two forms in Archaeplastida (plants and algae) is more complex. Plants exclusively possess the cobalamin-independent form, while algae have either one of the two, depending on species. Many different microorganisms express both the cobalamin-dependent and cobalamin-independent forms.
In enzymology, a leucocyanidin oxygenase (EC 1.14.11.19) is an enzyme that catalyzes the chemical reaction

The enzyme 3-dehydroquinate synthase catalyzes the chemical reaction
In enzymology, a 1-deoxy-d-xylulose-5-phosphate synthase (EC 2.2.1.7) is an enzyme in the non-mevalonate pathway that catalyzes the chemical reaction

Heme oxygenase 2 is an enzyme that in humans is encoded by the HMOX2 gene.
1-deoxy-11beta-hydroxypentalenate dehydrogenase (EC 1.1.1.340, 1-deoxy-11beta-hydroxypentalenic acid dehydrogenase, ptlF (gene), penF (gene name)) is an enzyme with systematic name 1-deoxy-11beta-hydroxypentalenate:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
Pentalenolactone synthase is an enzyme with systematic name pentalenolactone-F:oxidized-ferredoxin oxidoreductase . This enzyme catalyse the following chemical reaction
1-deoxypentalenic acid 11beta-hydroxylase (EC 1.14.11.35, PTLH (gene), SAV2991 (gene), PNTH (gene)) is an enzyme with systematic name 1-deoxypentalenic acid,2-oxoglutarate:oxygen oxidoreductase. This enzyme catalyses the following chemical reaction
Pentalenolactone F synthase (EC 1.14.11.36, PEND (gene), PNTD (gene), PTLD (gene)) is an enzyme with systematic name pentalenolactone-D,2-oxoglutarate:oxygen oxidoreductase. This enzyme catalyses the following chemical reaction
1-Deoxy-11-oxopentalenate,NADH:oxygen oxidoreductase may refer to:
Neopentalenolactone D synthase (EC 1.14.13.171, ptlE (gene)) is an enzyme with systematic name 1-deoxy-11-oxopentalenate,NADH:oxygen oxidoreductase (neopentalenolactone-D forming). This enzyme catalyses the following chemical reaction
Pentalenic acid synthase (EC 1.14.15.11, CYP105D7, sav7469 (gene)) is an enzyme with systematic name 1-deoxypentalenate,reduced ferredoxin:O2 oxidoreductase. This enzyme catalyses the following chemical reaction
6-deoxy-5-ketofructose 1-phosphate synthase is an enzyme with systematic name 2-oxopropanal:D-fructose 1,6-bisphosphate glycerone-phosphotransferase. This enzyme catalyses the following chemical reaction

Thiazole synthase (EC 2.8.1.10, thiG (gene)) is an enzyme with systematic name 1-deoxy-D-xylulose 5-phosphate:thiol sulfurtransferase. This enzyme catalyses the following chemical reaction
2-deoxy-scyllo-Inosose synthase is an enzyme with systematic name D-glucose-6-phosphate phosphate-lyase (2-deoxy-scyllo-inosose-forming). This enzyme catalyses the following chemical reaction
Alpha-ketoglutarate-dependent hydroxylases are a major class of non-heme iron proteins that catalyse a wide range of reactions. These reactions include hydroxylation reactions, demethylations, ring expansions, ring closures, and desaturations. Functionally, the αKG-dependent hydroxylases are comparable to cytochrome P450 enzymes. Both use O2 and reducing equivalents as cosubstrates and both generate water.
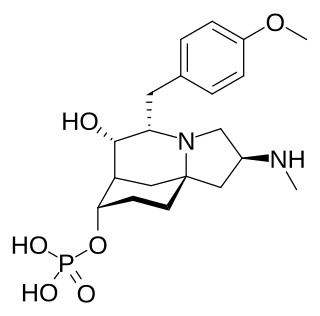
(−)-FR901483 is a tyrosine-derived alkaloid that was isolated from the fungus Cladobotryum sp. It was shown to have potent immunosuppressant activity in animal models. It is believed to function through inhibition of purine nucleotide biosynthesis.







