| Amino acid | Abbrev. | Remarks |
|---|
| Alanine | A | Ala | Very abundant and very versatile, it is more stiff than glycine, but small enough to pose only small steric limits for the protein conformation. It behaves fairly neutrally, and can be located in both hydrophilic regions on the protein outside and the hydrophobic areas inside. |
|---|
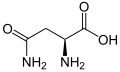
| Asparagine or aspartic acid | B | Asx | A placeholder when either amino acid may occupy a position |
|---|
| Cysteine | C | Cys | The sulfur atom bonds readily to heavy metal ions. Under oxidizing conditions, two cysteines can join in a disulfide bond to form the amino acid cystine. When cystines are part of a protein, insulin for example, the tertiary structure is stabilized, which makes the protein more resistant to denaturation; therefore, disulfide bonds are common in proteins that have to function in harsh environments including digestive enzymes (e.g., pepsin and chymotrypsin) and structural proteins (e.g., keratin). Disulfides are also found in peptides too small to hold a stable shape on their own (e.g. insulin). |
|---|
| Aspartic acid | D | Asp | Asp behaves similarly to glutamic acid, and carries a hydrophilic acidic group with strong negative charge. Usually, it is located on the outer surface of the protein, making it water-soluble. It binds to positively charged molecules and ions, and is often used in enzymes to fix the metal ion. When located inside of the protein, aspartate and glutamate are usually paired with arginine and lysine. |
|---|
| Glutamic acid | E | Glu | Glu behaves similarly to aspartic acid, and has a longer, slightly more flexible side chain. |
|---|
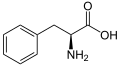
| Phenylalanine | F | Phe | Essential for humans, phenylalanine, tyrosine, and tryptophan contain a large, rigid aromatic group on the side chain. These are the biggest amino acids. Like isoleucine, leucine, and valine, these are hydrophobic and tend to orient towards the interior of the folded protein molecule. Phenylalanine can be converted into tyrosine. |
|---|
| Glycine | G | Gly | Because of the two hydrogen atoms at the α carbon, glycine is not optically active. It is the smallest amino acid, rotates easily, and adds flexibility to the protein chain. It is able to fit into the tightest spaces, e.g., the triple helix of collagen. As too much flexibility is usually not desired, as a structural component, it is less common than alanine. |
|---|
| Histidine | H | His | His is essential for humans. In even slightly acidic conditions, protonation of the nitrogen occurs, changing the properties of histidine and the polypeptide as a whole. It is used by many proteins as a regulatory mechanism, changing the conformation and behavior of the polypeptide in acidic regions such as the late endosome or lysosome, enforcing conformation change in enzymes. However, only a few histidines are needed for this, so it is comparatively scarce. |
|---|
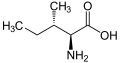
| Isoleucine | I | Ile | Ile is essential for humans. Isoleucine, leucine, and valine have large aliphatic hydrophobic side chains. Their molecules are rigid, and their mutual hydrophobic interactions are important for the correct folding of proteins, as these chains tend to be located inside of the protein molecule. |
|---|
| Leucine or isoleucine | J | Xle | A placeholder when either amino acid may occupy a position |
|---|
| Lysine | K | Lys | Lys is essential for humans, and behaves similarly to arginine. It contains a long, flexible side chain with a positively charged end. The flexibility of the chain makes lysine and arginine suitable for binding to molecules with many negative charges on their surfaces. E.g., DNA-binding proteins have their active regions rich with arginine and lysine. The strong charge makes these two amino acids prone to be located on the outer hydrophilic surfaces of the proteins; when they are found inside, they are usually paired with a corresponding negatively charged amino acid, e.g., aspartate or glutamate. |
|---|
| Leucine | L | Leu | Leu is essential for humans, and behaves similarly to isoleucine and valine. |
|---|
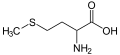
| Methionine | M | Met | Met is essential for humans. Always the first amino acid to be incorporated into a protein, it is sometimes removed after translation. Like cysteine, it contains sulfur, but with a methyl group instead of hydrogen. This methyl group can be activated, and is used in many reactions where a new carbon atom is being added to another molecule. |
|---|
| Asparagine | N | Asn | Similar to aspartic acid, Asn contains an amide group where Asp has a carboxyl. |
|---|
| Pyrrolysine | O | Pyl | Similar to lysine, but it has a pyrroline ring attached. |
|---|
| Proline | P | Pro | Pro contains an unusual ring to the N-end amine group, which forces the CO-NH amide sequence into a fixed conformation. It can disrupt protein folding structures like α helix or β sheet, forcing the desired kink in the protein chain. Common in collagen, it often undergoes a post-translational modification to hydroxyproline. |
|---|
| Glutamine | Q | Gln | Similar to glutamic acid, Gln contains an amide group where Glu has a carboxyl. Used in proteins and as a storage for ammonia, it is the most abundant amino acid in the body. |
|---|
| Arginine | R | Arg | Functionally similar to lysine. |
|---|
| Serine | S | Ser | Serine and threonine have a short group ended with a hydroxyl group. Its hydrogen is easy to remove, so serine and threonine often act as hydrogen donors in enzymes. Both are very hydrophilic, so the outer regions of soluble proteins tend to be rich with them. |
|---|
| Threonine | T | Thr | Essential for humans, Thr behaves similarly to serine. |
|---|
| Selenocysteine | U | Sec | The selenium analog of cysteine, in which selenium replaces the sulfur atom. |
|---|
| Valine | V | Val | Essential for humans, Val behaves similarly to isoleucine and leucine. |
|---|
| Tryptophan | W | Trp | Essential for humans, Trp behaves similarly to phenylalanine and tyrosine. It is a precursor of serotonin and is naturally fluorescent. |
|---|
| Unknown | X | Xaa | Placeholder when the amino acid is unknown or unimportant. |
|---|
| Tyrosine | Y | Tyr | Tyr behaves similarly to phenylalanine (precursor to tyrosine) and tryptophan, and is a precursor of melanin, epinephrine, and thyroid hormones. Naturally fluorescent, its fluorescence is usually quenched by energy transfer to tryptophans. |
|---|
| Glutamic acid or glutamine | Z | Glx | A placeholder when either amino acid may occupy a position |
|---|