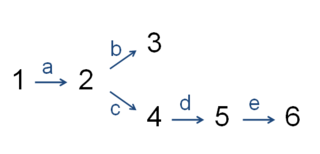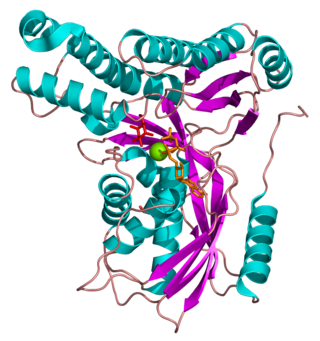
Galactokinase is an enzyme (phosphotransferase) that facilitates the phosphorylation of α-D-galactose to galactose 1-phosphate at the expense of one molecule of ATP. Galactokinase catalyzes the second step of the Leloir pathway, a metabolic pathway found in most organisms for the catabolism of α-D-galactose to glucose 1-phosphate. First isolated from mammalian liver, galactokinase has been studied extensively in yeast, archaea, plants, and humans.

Polymyxins are antibiotics. Polymyxins B and E are used in the treatment of Gram-negative bacterial infections. They work mostly by breaking up the bacterial cell membrane. They are part of a broader class of molecules called nonribosomal peptides.

Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.
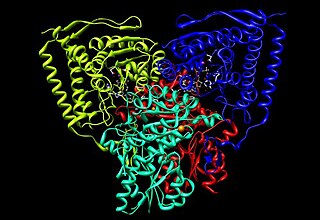
Pyruvate dehydrogenase is an enzyme that catalyzes the reaction of pyruvate and a lipoamide to give the acetylated dihydrolipoamide and carbon dioxide. The conversion requires the coenzyme thiamine pyrophosphate.

The enzyme Acid-Induced Arginine Decarboxylase (AdiA), also commonly referred to as arginine decarboxylase, catalyzes the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase. It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH. Arginine decarboxylase is part of an enzymatic system in Escherichia coli, Salmonella Typhimurium, and methane-producing bacteria Methanococcus jannaschii that makes these organisms acid resistant and allows them to survive under highly acidic medium.

The enzyme diaminopimelate decarboxylase (EC 4.1.1.20) catalyzes the cleavage of carbon-carbon bonds in meso 2,6 diaminoheptanedioate to produce CO2 and L-lysine, the essential amino acid. It employs the cofactor pyridoxal phosphate, also known as PLP, which participates in numerous enzymatic transamination, decarboxylation and deamination reactions.
In enzymology, a N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase is an enzyme that catalyzes the chemical reaction
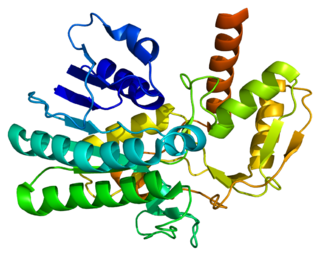
UDP-glucuronic acid decarboxylase 1 is an enzyme that in humans is encoded by the UXS1 gene.
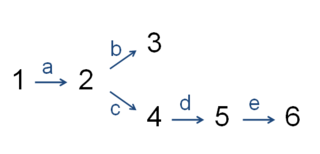
In enzymology, the committed step is an effectively irreversible enzymatic reaction that occurs at a branch point during the biosynthesis of some molecules. As the name implies, after this step, the molecules are "committed" to the pathway and will ultimately end up in the pathway's final product. The first committed step should not be confused with the rate-limiting step, which is the step with the highest flux control coefficient. It is rare that the first committed step is in fact the rate-determining step.
David Sidney Feingold was an American biochemist.
In molecular biology, the lipopolysaccharide kinase (Kdo/WaaP) family is a family of lipopolysaccharide kinases that includes lipopolysaccharide core heptose(I) kinase rfaP. Lipopolysaccharide core heptose(I) kinase rfaP is required for the addition of phosphate to O-4 of the first heptose residue of the lipopolysaccharide (LPS) inner core region. It has previously been shown that it is necessary for resistance to hydrophobic and polycationic antimicrobials in E. coli and that it is required for virulence in invasive strains of Salmonella enterica. The family also includes 3-deoxy-D-manno-octulosonic acid kinase from Haemophilus influenzae, which phosphorylates Kdo-lipid IV(A), a lipopolysaccharide precursor, and is involved in virulence.
UDP-N-acetyl-2-amino-2-deoxyglucuronate dehydrogenase (EC 1.1.1.335, WlbA, WbpB) is an enzyme with systematic name UDP-N-acetyl-2-amino-2-deoxy-alpha-D-glucuronate:NAD+ 3-oxidoreductase. This enzyme catalyses the following chemical reaction:
UDP-4-amino-4-deoxy-L-arabinose formyltransferase is an enzyme with systematic name 10-formyltetrahydrofolate:UDP-4-amino-4-deoxy-beta-L-arabinose N-formyltransferase. This enzyme catalyses the following chemical reaction
Cyanidin-3-O-glucoside 2-O-glucuronosyltransferase is an enzyme with systematic name UDP-D-glucuronate:cyanidin-3-O-beta-D-glucoside 2-O-beta-D-glucuronosyltransferase. This enzyme catalyses the following chemical reaction
Lipid IVA 4-amino-4-deoxy-L-arabinosyltransferase is an enzyme with systematic name 4-amino-4-deoxy-alpha-L-arabinopyranosyl ditrans, octacis-undecaprenyl phosphate:lipid IVA 4-amino-4-deoxy-L-arabinopyranosyltransferase. This enzyme catalyses the following chemical reaction
Lipid IVA 3-deoxy-D-manno-octulosonic acid transferase is an enzyme with systematic name CMP-3-deoxy-D-manno-oct-2-ulosonate:lipid IVA 3-deoxy-D-manno-oct-2-ulosonate transferase. This enzyme catalyses the following chemical reaction
UDP-4-amino-4-deoxy-L-arabinose aminotransferase is an enzyme with systematic name UDP-4-amino-4-deoxy-beta-L-arabinose:2-oxoglutarate aminotransferase. This enzyme catalyses the following chemical reaction
UDP-2-acetamido-2-deoxy-ribo-hexuluronate aminotransferase is an enzyme with systematic name UDP-2-acetamido-3-amino-2,3-dideoxy-alpha-D-glucuronate:2-oxoglutarate aminotransferase. This enzyme catalyses the following chemical reaction
3-deoxy-D-manno-octulosonic acid kinase is an enzyme with systematic name ATP:(KDO)-lipid IVA 3-deoxy-alpha-D-manno-oct-2-ulopyranose 4-phosphotransferase. This enzyme catalyses the following chemical reaction
Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase is an enzyme with systematic name UDP-4-amino-4-deoxy-alpha-L-arabinose:ditrans,octacis-undecaprenyl phosphate 4-amino-4-deoxy-alpha-L-arabinosyltransferase. This enzyme catalyses the following chemical reaction