| cyclohexadienyl dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC no. | 1.3.1.43 | ||||||||
| CAS no. | 64295-75-6 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
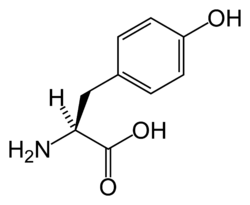
In enzymology, arogenate dehydrogenase (EC 1.3.1.43) is an enzyme that catalyzes the chemical reaction
The two substrates of this enzyme are L-arogenic acid (shown as its conjugate base arogenate) and oxidised nicotinamide adenine dinucleotide (NAD+). Its products are L-tyrosine, reduced NADH, and carbon dioxide. [1] [2] [3]
This enzyme belongs to the family of oxidoreductases, specifically those acting on the CH-CH group of donor with NAD+ or NADP+ as acceptor. The systematic name of this enzyme class is L-arogenate:NAD+ oxidoreductase (decarboxylating). Other names in common use include arogenic dehydrogenase (ambiguous), cyclohexadienyl dehydrogenase, pretyrosine dehydrogenase (ambiguous), and L-arogenate:NAD+ oxidoreductase. This enzyme participates in phenylalanine, tyrosine and tryptophan biosynthesis and novobiocin biosynthesis. [4] [5] [6]