The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a specific gene depends on the number of copies of each chromosome found in that species, also referred to as ploidy. In diploid species like humans, two full sets of chromosomes are present, meaning each individual has two alleles for any given gene. If both alleles are the same, the genotype is referred to as homozygous. If the alleles are different, the genotype is referred to as heterozygous.

Mendelian inheritance is a type of biological inheritance that follows the principles originally proposed by Gregor Mendel in 1865 and 1866, re-discovered in 1900 by Hugo de Vries and Carl Correns, and popularized by William Bateson. These principles were initially controversial. When Mendel's theories were integrated with the Boveri–Sutton chromosome theory of inheritance by Thomas Hunt Morgan in 1915, they became the core of classical genetics. Ronald Fisher combined these ideas with the theory of natural selection in his 1930 book The Genetical Theory of Natural Selection, putting evolution onto a mathematical footing and forming the basis for population genetics within the modern evolutionary synthesis.

In biology, a hybrid is the offspring resulting from combining the qualities of two organisms of different breeds, varieties, species or genera through sexual reproduction. Hybrids are not always intermediates between their parents, but can show hybrid vigor, sometimes growing larger or taller than either parent. The concept of a hybrid is interpreted differently in animal and plant breeding, where there is interest in the individual parentage. In genetics, attention is focused on the numbers of chromosomes. In taxonomy, a key question is how closely related the parent species are.

In genetics, dominance is the phenomenon of one variant (allele) of a gene on a chromosome masking or overriding the effect of a different variant of the same gene on the other copy of the chromosome. The first variant is termed dominant and the second recessive. This state of having two different variants of the same gene on each chromosome is originally caused by a mutation in one of the genes, either new or inherited. The terms autosomal dominant or autosomal recessive are used to describe gene variants on non-sex chromosomes (autosomes) and their associated traits, while those on sex chromosomes (allosomes) are termed X-linked dominant, X-linked recessive or Y-linked; these have an inheritance and presentation pattern that depends on the sex of both the parent and the child. Since there is only one copy of the Y chromosome, Y-linked traits cannot be dominant or recessive. Additionally, there are other forms of dominance such as incomplete dominance, in which a gene variant has a partial effect compared to when it is present on both chromosomes, and co-dominance, in which different variants on each chromosome both show their associated traits.
Genetic linkage is the tendency of DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two genetic markers that are physically near to each other are unlikely to be separated onto different chromatids during chromosomal crossover, and are therefore said to be more linked than markers that are far apart. In other words, the nearer two genes are on a chromosome, the lower the chance of recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly unlinked, although the penetrance of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located.
Inbred strains are individuals of a particular species which are nearly identical to each other in genotype due to long inbreeding. A strain is inbred when it has undergone at least 20 generations of brother x sister or offspring x parent mating, at which point at least 98.6% of the loci in an individual of the strain will be homozygous, and each individual can be treated effectively as clones. Some inbred strains have been bred for over 150 generations, leaving individuals in the population to be isogenic in nature. Inbred strains of animals are frequently used in laboratories for experiments where for the reproducibility of conclusions all the test animals should be as similar as possible. However, for some experiments, genetic diversity in the test population may be desired. Thus outbred strains of most laboratory animals are also available, where an outbred strain is a strain of an organism that is effectively wildtype in nature, where there is as little inbreeding as possible.
Heterosis, hybrid vigor, or outbreeding enhancement is the improved or increased function of any biological quality in a hybrid offspring. An offspring is heterotic if its traits are enhanced as a result of mixing the genetic contributions of its parents. These effects can be due to Mendelian or non-Mendelian inheritance.

A crossbreed is an organism with purebred parents of two different breeds, varieties, or populations. Crossbreeding, sometimes called "designer crossbreeding", is the process of breeding such an organism, While crossbreeding is used to maintain health and viability of organisms, irresponsible crossbreeding can also produce organisms of inferior quality or dilute a purebred gene pool to the point of extinction of a given breed of organism.
An F1 hybrid (also known as filial 1 hybrid) is the first filial generation of offspring of distinctly different parental types. F1 hybrids are used in genetics, and in selective breeding, where the term F1 crossbreed may be used. The term also is sometimes written with a subscript, as F1 hybrid. Subsequent generations are called F2, F3, etc.

"Open pollination" and "open pollinated" refer to a variety of concepts in the context of the sexual reproduction of plants. Generally speaking, the term refers to plants pollinated naturally by birds, insects, wind, or human hands.
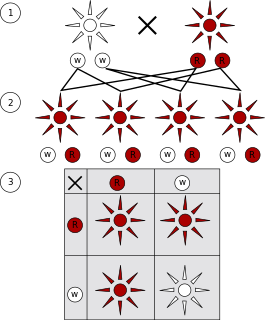
A monohybrid cross is a cross between two organisms with different variations at one genetic locus of interest. The character(s) being studied in a monohybrid cross are governed by two or multiple variations for a single location of a gene. To carry out such a cross, each parent is chosen to be homozygous or true breeding for a given trait (locus). When a cross satisfies the conditions for a monohybrid cross, it is usually detected by a characteristic distribution of second-generation (F2) offspring that is sometimes called the monohybrid ratio.
Out-crossing or out-breeding is the technique of crossing between different breeds. This is the practice of introducing distantly related genetic material into a breeding line, thereby increasing genetic diversity.
In genetics, two organisms that differ in only one locus and a linked segment of chromosome are defined as congenic. Similarly, organisms that are coisogenic differ in one locus only and not in the surrounding chromosome. Unlike congenic organisms, coisogenic organisms cannot be bred and only occur through spontaneous or targeted mutation at the locus.
Marker assisted selection or marker aided selection (MAS) is an indirect selection process where a trait of interest is selected based on a marker linked to a trait of interest, rather than on the trait itself. This process has been extensively researched and proposed for plant and animal breeding.
Senecio eboracensis, the York groundsel or York radiate groundsel, is a flowering plant in the daisy family Asteraceae. It is a self-pollinating hybrid species of ragwort and one of only six new plant species to be discovered in either the United Kingdom or North America in the last 100 years. It was discovered in 1979 in York, England growing next to a car park and formally described in 2003. Like many of the Senecio genus it can be found growing in urban habitats, such as disturbed earth and pavement cracks and this particular species only in York and between a railway and a car park.
A doubled haploid (DH) is a genotype formed when haploid cells undergo chromosome doubling. Artificial production of doubled haploids is important in plant breeding.

Plant breeding is the science of changing the traits of plants in order to produce desired characteristics. It has been used to improve the quality of nutrition in products for humans and animals. The goals of plant breeding are to produce crop varieties that boast unique and superior traits for a variety of agricultural applications. The most frequently addressed traits are those related to biotic and abiotic stress tolerance, grain or biomass yield, end-use quality characteristics such as taste or the concentrations of specific biological molecules and ease of processing.
A knockout mouse, or knock-out mouse, is a genetically modified mouse in which researchers have inactivated, or "knocked out", an existing gene by replacing it or disrupting it with an artificial piece of DNA. They are important animal models for studying the role of genes which have been sequenced but whose functions have not been determined. By causing a specific gene to be inactive in the mouse, and observing any differences from normal behaviour or physiology, researchers can infer its probable function.
Molecular breeding is the application of molecular biology tools, often in plant breeding and animal breeding. In the broad sense, molecular breeding can be defined as the use of genetic manipulation performed at the level of DNA to improve traits of interest in plants and animals, and it may also include genetic engineering or gene manipulation, molecular marker-assisted selection, and genomic selection. More often, however, molecular breeding implies molecular marker-assisted breeding (MAB) and is defined as the application of molecular biotechnologies, specifically molecular markers, in combination with linkage maps and genomics, to alter and improve plant or animal traits on the basis of genotypic assays.
Classical genetics is the branch of genetics based solely on visible results of reproductive acts. It is the oldest discipline in the field of genetics, going back to the experiments on Mendelian inheritance by Gregor Mendel who made it possible to identify the basic mechanisms of heredity. Subsequently, these mechanisms have been studied and explained at the molecular level.








