
Enterococcus is a large genus of lactic acid bacteria of the phylum Bacillota. Enterococci are Gram-positive cocci that often occur in pairs (diplococci) or short chains, and are difficult to distinguish from streptococci on physical characteristics alone. Two species are common commensal organisms in the intestines of humans: E. faecalis (90–95%) and E. faecium (5–10%). Rare clusters of infections occur with other species, including E. casseliflavus, E. gallinarum, and E. raffinosus.

An agar plate is a Petri dish that contains a growth medium solidified with agar, used to culture microorganisms. Sometimes selective compounds are added to influence growth, such as antibiotics.

Bacteriological water analysis is a method of analysing water to estimate the numbers of bacteria present and, if needed, to find out what sort of bacteria they are. It represents one aspect of water quality. It is a microbiological analytical procedure which uses samples of water and from these samples determines the concentration of bacteria. It is then possible to draw inferences about the suitability of the water for use from these concentrations. This process is used, for example, to routinely confirm that water is safe for human consumption or that bathing and recreational waters are safe to use.

Coliform bacteria are defined as either motile or non-motile Gram-negative non-spore forming bacilli that possess β-galactosidase to produce acids and gases under their optimal growth temperature of 35–37 °C. They can be aerobes or facultative aerobes, and are a commonly used indicator of low sanitary quality of foods, milk, and water. Coliforms can be found in the aquatic environment, in soil and on vegetation; they are universally present in large numbers in the feces of warm-blooded animals as they are known to inhabit the gastrointestinal system. While coliform bacteria are not normally the cause of serious illness, they are easy to culture, and their presence is used to infer that other pathogenic organisms of fecal origin may be present in a sample, or that said sample is not safe to consume. Such pathogens include disease-causing bacteria, viruses, or protozoa and many multicellular parasites. Every drinking water source must be tested for the presence of these total coliform bacteria.

MacConkey agar is a selective and differential culture medium for bacteria. It is designed to selectively isolate gram-negative and enteric bacteria and differentiate them based on lactose fermentation. Lactose fermenters turn red or pink on MacConkey agar, and nonfermenters do not change color. The media inhibits growth of gram-positive organisms with crystal violet and bile salts, allowing for the selection and isolation of gram-negative bacteria. The media detects lactose fermentation by enteric bacteria with the pH indicator neutral red.
Bartonellosis is an infectious disease produced by bacteria of the genus Bartonella. Bartonella species cause diseases such as Carrión's disease, trench fever, cat-scratch disease, bacillary angiomatosis, peliosis hepatis, chronic bacteremia, endocarditis, chronic lymphadenopathy, and neurological disorders.
Sorbitol-MacConkey agar is a variant of traditional MacConkey agar used in the detection of E. coli O157:H7. Traditionally, MacConkey agar has been used to distinguish those bacteria that ferment lactose from those that do not. This is important because gut bacteria, such as Escherichia coli, can typically ferment lactose, while important gut pathogens, such as Salmonella enterica and most shigellas are unable to ferment lactose. Shigella sonnei can ferment lactose, but only after prolonged incubation, so it is referred to as a late-lactose fermenter.

Burkholderia pseudomallei is a Gram-negative, bipolar, aerobic, motile rod-shaped bacterium. It is a soil-dwelling bacterium endemic in tropical and subtropical regions worldwide, particularly in Thailand and northern Australia. It was reported in 2008 that there had been an expansion of the affected regions due to significant natural disasters, and it could be found in Southern China, Hong Kong, and countries in the Americas. B. pseudomallei, amongst other pathogens, has been found in monkeys imported into the United States from Asia for laboratory use, posing a risk that the pathogen could be introduced into the country.
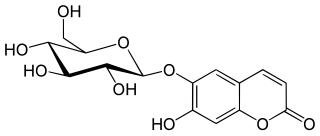
Aesculin, also called æsculin or esculin, is a coumarin glucoside that naturally occurs in the trees horse chestnut, California buckeye, prickly box, and daphnin. It is also found in dandelion coffee and olive bark. It is reported to present in olive bark, not in olive leaf, therefore, identification of aesculin in abundant in an extract indicates the extract derived from olive bark.
Mycobacterium bohemicum is a species of the phylum Actinomycetota, belonging to the genus Mycobacterium.
Mycobacterium farcinogenes is a species of the phylum Actinomycetota, belonging to the genus Mycobacterium.

Ashdown's medium is a selective culture medium for the isolation and characterisation of Burkholderia pseudomallei.

Hektoen enteric agar is a selective and differential agar primarily used to recover Salmonella and Shigella from patient specimens. HEA contains indicators of lactose fermentation and hydrogen sulfide production; as well as inhibitors to prevent the growth of Gram-positive bacteria. It is named after the Hektoen Institute in Chicago, where researchers developed the agar.

Thiosulfate–citrate–bile salts–sucrose agar, or TCBS agar, is a type of selective agar culture plate that is used in microbiology laboratories to isolate Vibrio species. TCBS agar is highly selective for the isolation of V. cholerae and V. parahaemolyticus as well as other Vibrio species. Apart from TCBS agar, other rapid testing dipsticks like immunochromatographic dipstick is also used in endemic areas such as Asia, Africa and Latin America. Though, TCBS agar study is required for confirmation. This becomes immensely important in cases of gastroenteritis caused by Campylobacter species, whose symptoms mimic those of cholera. Since no yellow bacterial growth is observed in case of Campylobacter species on TCBS agar, chances of incorrect diagnosis can be rectified. TCBS agar contains high concentrations of sodium thiosulfate and sodium citrate to inhibit the growth of Enterobacteriaceae. Inhibition of gram-positive bacteria is achieved by the incorporation of ox gall, which is a naturally occurring substance containing a mixture of bile salts and sodium cholate, a pure bile salt. Sodium thiosulfate also serves as a sulfur source and its presence, in combination with ferric citrate, allows for the easy detection of hydrogen sulfide production. Saccharose (sucrose) is included as a fermentable carbohydrate for metabolism by Vibrio species. The alkaline pH of the medium enhances the recovery of V. cholerae and inhibits the growth of others. Thymol blue and bromothymol blue are included as indicators of pH changes.

Proteus penneri is a Gram-negative, facultatively anaerobic, rod-shaped bacterium. It is an invasive pathogen and a cause of nosocomial infections of the urinary tract or open wounds. Pathogens have been isolated mainly from the urine of patients with abnormalities in the urinary tract, and from stool. P. penneri strains are naturally resistant to numerous antibiotics, including penicillin G, amoxicillin, cephalosporins, oxacillin, and most macrolides, but are naturally sensitive to aminoglycosides, carbapenems, aztreonam, quinolones, sulphamethoxazole, and co-trimoxazole. Isolates of P. penneri have been found to be multiple drug-resistant (MDR) with resistance to six to eight drugs. β-lactamase production has also been identified in some isolates.
Enterococcus malodoratus is a species of the genus Enterococcus and a gram positive bacteria capable of opportunistic pathogenic response. These microbes have a thick polypeptide layer. Enterococcus can be found in the gastrointestinal tracts of humans and other mammals. In a study on the enterococcal flora of swine, E. malodoratus was found in the intestines and feces. It was not identified within the tonsils of swine, nor within cats, calves, dogs, horse, or poultry. The name "malodoratus" translates to "ill smelling".

Granada medium is a selective and differential culture medium designed to selectively isolate Streptococcus agalactiae and differentiate it from other microorganisms. Granada Medium was developed by Manuel Rosa-Fraile et al. at the Service of Microbiology in the Hospital Virgen de las Nieves in Granada (Spain).
Diagnostic microbiology is the study of microbial identification. Since the discovery of the germ theory of disease, scientists have been finding ways to harvest specific organisms. Using methods such as differential media or genome sequencing, physicians and scientists can observe novel functions in organisms for more effective and accurate diagnosis of organisms. Methods used in diagnostic microbiology are often used to take advantage of a particular difference in organisms and attain information about what species it can be identified as, which is often through a reference of previous studies. New studies provide information that others can reference so that scientists can attain a basic understanding of the organism they are examining.
Columbia Nalidixic Acid (CNA) agar is a growth medium used for the isolation and cultivation of bacteria from clinical and non-clinical specimens. CNA agar contains antibiotics that inhibit Gram-negative organisms, aiding in the selective isolation of Gram-positive bacteria. Gram-positive organisms that grow on the media can be differentiated on the basis of hemolysis.
Enterococcus casseliflavus is a species of commensal Gram-positive bacteria. Its name derived from the "flavus" the Latin word for yellow due to the bright yellow pigment that it produces. This organism can be found in the gastrointestinal tract of humans












