Cell biology is a branch of biology that studies the structure, function, and behavior of cells. All living organisms are made of cells. A cell is the basic unit of life that is responsible for the living and functioning of organisms. Cell biology is the study of the structural and functional units of cells. Cell biology encompasses both prokaryotic and eukaryotic cells and has many subtopics which may include the study of cell metabolism, cell communication, cell cycle, biochemistry, and cell composition. The study of cells is performed using several microscopy techniques, cell culture, and cell fractionation. These have allowed for and are currently being used for discoveries and research pertaining to how cells function, ultimately giving insight into understanding larger organisms. Knowing the components of cells and how cells work is fundamental to all biological sciences while also being essential for research in biomedical fields such as cancer, and other diseases. Research in cell biology is interconnected to other fields such as genetics, molecular genetics, molecular biology, medical microbiology, immunology, and cytochemistry.

A mitochondrion is an organelle found in the cells of most eukaryotes, such as animals, plants and fungi. Mitochondria have a double membrane structure and use aerobic respiration to generate adenosine triphosphate (ATP), which is used throughout the cell as a source of chemical energy. They were discovered by Albert von Kölliker in 1857 in the voluntary muscles of insects. The term mitochondrion was coined by Carl Benda in 1898. The mitochondrion is popularly nicknamed the "powerhouse of the cell", a phrase coined by Philip Siekevitz in a 1957 article of the same name.
A proton pump is an integral membrane protein pump that builds up a proton gradient across a biological membrane. Proton pumps catalyze the following reaction:

ATP synthase is a protein that catalyzes the formation of the energy storage molecule adenosine triphosphate (ATP) using adenosine diphosphate (ADP) and inorganic phosphate (Pi). ATP synthase is a molecular machine. The overall reaction catalyzed by ATP synthase is:

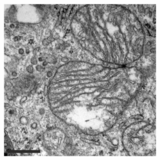
A crista is a fold in the inner membrane of a mitochondrion. The name is from the Latin for crest or plume, and it gives the inner membrane its characteristic wrinkled shape, providing a large amount of surface area for chemical reactions to occur on. This aids aerobic cellular respiration, because the mitochondrion requires oxygen. Cristae are studded with proteins, including ATP synthase and a variety of cytochromes.

The inner mitochondrial membrane (IMM) is the mitochondrial membrane which separates the mitochondrial matrix from the intermembrane space.

Mitochondrial membrane transport proteins, also known as mitochondrial carrier proteins, are proteins which exist in the membranes of mitochondria. They serve to transport molecules and other factors, such as ions, into or out of the organelles. Mitochondria contain both an inner and outer membrane, separated by the inter-membrane space, or inner boundary membrane. The outer membrane is porous, whereas the inner membrane restricts the movement of all molecules. The two membranes also vary in membrane potential and pH. These factors play a role in the function of mitochondrial membrane transport proteins. There are 53 discovered human mitochondrial membrane transporters, with many others that are known to still need discovered.

Mitofusin-2 is a protein that in humans is encoded by the MFN2 gene. Mitofusins are GTPases embedded in the outer membrane of the mitochondria. In mammals MFN1 and MFN2 are essential for mitochondrial fusion. In addition to the mitofusins, OPA1 regulates inner mitochondrial membrane fusion, and DRP1 is responsible for mitochondrial fission.

Dynamin-1-like protein is a GTPase that regulates mitochondrial fission. In humans, dynamin-1-like protein, which is typically referred to as dynamin-related protein 1 (Drp1), is encoded by the DNM1L gene and is part of the dynamin superfamily (DSP) family of proteins.

Mitochondrial fission 1 protein (FIS1) is a protein that in humans is encoded by the FIS1 gene on chromosome 7. This protein is a component of a mitochondrial complex, the ARCosome, that promotes mitochondrial fission. Its role in mitochondrial fission thus implicates it in the regulation of mitochondrial morphology, the cell cycle, and apoptosis. By extension, the protein is involved in associated diseases, including neurodegenerative diseases and cancers.

ATP-dependent metalloprotease YME1L1 is an enzyme that in humans is encoded by the YME1L1 gene. YME1L1 belongs to the AAA family of ATPases and mainly functions in the maintenance of mitochondrial morphology. Mutations in this gene would cause infantile-onset mitochondriopathy.
Mitochondrial biogenesis is the process by which cells increase mitochondrial numbers. It was first described by John Holloszy in the 1960s, when it was discovered that physical endurance training induced higher mitochondrial content levels, leading to greater glucose uptake by muscles. Mitochondrial biogenesis is activated by numerous different signals during times of cellular stress or in response to environmental stimuli, such as aerobic exercise.
Fission, in biology, is the division of a single entity into two or more parts and the regeneration of those parts to separate entities resembling the original. The object experiencing fission is usually a cell, but the term may also refer to how organisms, bodies, populations, or species split into discrete parts. The fission may be binary fission, in which a single organism produces two parts, or multiple fission, in which a single entity produces multiple parts.
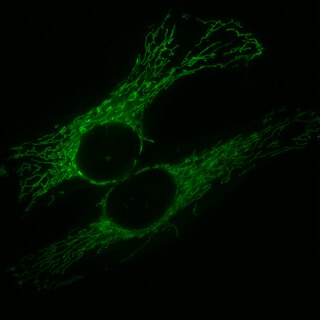
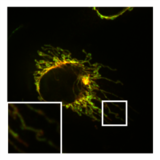
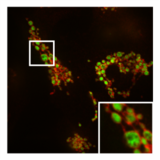
Mitochondria are dynamic organelles with the ability to fuse and divide (fission), forming constantly changing tubular networks in most eukaryotic cells. These mitochondrial dynamics, first observed over a hundred years ago are important for the health of the cell, and defects in dynamics lead to genetic disorders. Through fusion, mitochondria can overcome the dangerous consequences of genetic malfunction. The process of mitochondrial fusion involves a variety of proteins that assist the cell throughout the series of events that form this process.

The phomoxanthones are a loosely defined class of natural products. The two founding members of this class are phomoxanthone A and phomoxanthone B. Other compounds were later also classified as phomoxanthones, although a unifying nomenclature has not yet been established. The structure of all phomoxanthones is derived from a dimer of two covalently linked tetrahydroxanthones, and they differ mainly in the position of this link as well as in the acetylation status of their hydroxy groups. The phomoxanthones are structurally closely related to other tetrahydroxanthone dimers such as the secalonic acids and the eumitrins. While most phomoxanthones were discovered in fungi of the genus Phomopsis, most notably in the species Phomopsis longicolla, some have also been found in Penicillium sp.
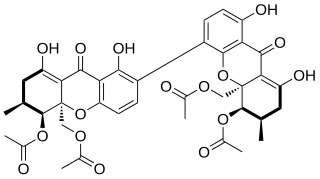
The mycotoxin phomoxanthone B, or PXB for short, is a toxic natural product. It is a less toxic isomer of phomoxanthone A and one of the two founding members of the class of phomoxanthone compounds. The phomoxanthones are named after the fungus Phomopsis, from which they were first isolated, and after their xanthonoid structure. Chemically, they are dimers of two tetrahydroxanthones that are covalently linked to each other. PXB itself is a homodimer of two identical diacetylated tetrahydroxanthones. The position of the link between the two tetrahydroxanthones is the only structural difference between PXB and its isomers PXA and dicerandrol C: In PXA, the two xanthonoid monomers are symmetrically linked at C-4,4’, while in PXB, they are asymmetrically linked at C-2,4’, and in dicerandrol C, they are symmetrically linked at C-2,2’.
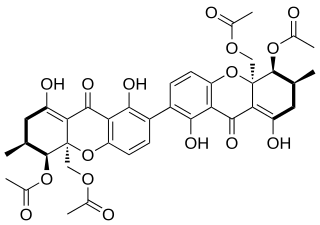
Dicerandrol C is a natural product. It is a less toxic isomer of phomoxanthone A (PXA) and phomoxanthone B (PXB), all three of which are members of the class of phomoxanthone compounds. The phomoxanthones are named after the fungus Phomopsis, from which they were first isolated, and after their xanthonoid structure. Chemically, they are dimers of two tetrahydroxanthones that are covalently linked to each other. Dicerandrol C itself is a homodimer of two identical diacetylated tetrahydroxanthones. The position of the link between the two tetrahydroxanthones is the only structural difference between dicerandrol C and its isomers PXA and PXB: In PXA, the two xanthonoid monomers are symmetrically linked at C-4,4’, while in PXB, they are asymmetrically linked at C-2,4’, and in dicerandrol C, they are symmetrically linked at C-2,2’.

Mitochondrial elongation factor 2 is a protein that in humans is encoded by the MIEF2 gene.

GFER Syndrome is a rare mitochondrial disease. GFER was first reported in 2009 and since exome sequencing became more available, few more cases were discovered. In all known cases, the disease progresses with conditions that include: congenital cataracts, loss of motor abilities, development delay, degeneration of organs, sometimes hearing loss, etc.

Solute carrier family 25 member 46 is a protein that in humans is encoded by the SLC25A46 gene. This protein is a member of the SLC25 mitochondrial solute carrier family. It is a transmembrane protein located in the mitochondrial outer membrane involved in lipid transfer from the endoplasmic reticulum (ER) to mitochondria. Mutations in this gene result in neuropathy and optic atrophy.