Related Research Articles

Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion.

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.
A metalloproteinase, or metalloprotease, is any protease enzyme whose catalytic mechanism involves a metal. An example is ADAM12 which plays a significant role in the fusion of muscle cells during embryo development, in a process known as myogenesis.

Transglutaminases are enzymes that in nature primarily catalyze the formation of an isopeptide bond between γ-carboxamide groups ( -(C=O)NH2 ) of glutamine residue side chains and the ε-amino groups ( -NH2 ) of lysine residue side chains with subsequent release of ammonia ( NH3 ). Lysine and glutamine residues must be bound to a peptide or a protein so that this cross-linking (between separate molecules) or intramolecular (within the same molecule) reaction can happen. Bonds formed by transglutaminase exhibit high resistance to proteolytic degradation (proteolysis). The reaction is

Dihydrolipoamide dehydrogenase (DLD), also known as dihydrolipoyl dehydrogenase, mitochondrial, is an enzyme that in humans is encoded by the DLD gene. DLD is a flavoprotein enzyme that oxidizes dihydrolipoamide to lipoamide.

Insulin-degrading enzyme, also known as IDE, is an enzyme.
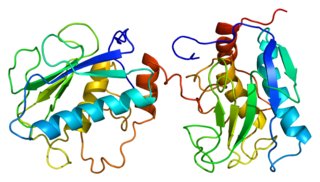
Stromelysin-1 also known as matrix metalloproteinase-3 (MMP-3) is an enzyme that in humans is encoded by the MMP3 gene. The MMP3 gene is part of a cluster of MMP genes which localize to chromosome 11q22.3. MMP-3 has an estimated molecular weight of 54 kDa.

Matrilysin also known as matrix metalloproteinase-7 (MMP-7), pump-1 protease (PUMP-1), or uterine metalloproteinase is an enzyme in humans that is encoded by the MMP7 gene. The enzyme has also been known as matrin, putative metalloproteinase-1, matrix metalloproteinase pump 1, PUMP-1 proteinase, PUMP, metalloproteinase pump-1, putative metalloproteinase, MMP). Human MMP-7 has a molecular weight around 30 kDa.
The discovery of an orally inactive peptide from snake venom established the important role of angiotensin converting enzyme (ACE) inhibitors in regulating blood pressure. This led to the development of captopril, the first ACE inhibitor. When the adverse effects of captopril became apparent new derivates were designed. Then after the discovery of two active sites of ACE: N-domain and C-domain, the development of domain-specific ACE inhibitors began.
In enzymology, an aminoacylase (EC 3.5.1.14) is an enzyme that catalyzes the chemical reaction

A Disintegrin and metalloproteinase domain-containing protein 10, also known as ADAM10 or CDw156 or CD156c is a protein that in humans is encoded by the ADAM10 gene.

Dipeptidyl-peptidase 3 is an enzyme that in humans is encoded by the DPP3 gene.
Dipeptidyl-peptidase III is an enzyme. This enzyme catalyses the following chemical reaction
Dihydroorotate dehydrogenase (fumarate) (EC 1.3.98.1, dihydroorotate oxidase, pyr4 (gene)) is an enzyme with systematic name (S)-dihydroorotate:fumarate oxidoreductase. This enzyme catalyses the following chemical reaction
Aminopeptidase S is an enzyme. This enzyme catalyses the following chemical reaction
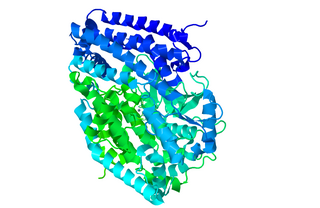
Peptidyl-dipeptidase Dcp (EC 3.4.15.5, dipeptidyl carboxypeptidase (Dcp), dipeptidyl carboxypeptidase) is a metalloenzyme found in the cytoplasm of bacterium E. Coli responsible for the C-terminal cleavage of a variety of dipeptides and unprotected larger peptide chains. The enzyme does not hydrolyze bonds in which P1' is Proline, or both P1 and P1' are Glycine. Dcp consists of 680 amino acid residues that form into a single active monomer which aids in the intracellular degradation of peptides. Dcp coordinates to divalent zinc which sits in the pocket of the active site and is composed of four subsites: S1’, S1, S2, and S3, each subsite attracts certain amino acids at a specific position on the substrate enhancing the selectivity of the enzyme. The four subsites detect and bind different amino acid types on the substrate peptide in the P1 and P2 positions. Some metallic divalent cations such as Ni+2, Cu+2, and Zn+2 inhibit the function of the enzyme around 90%, whereas other cations such as Mn+2, Ca+2, Mg+2, and Co+2 have slight catalyzing properties, and increase the function by around 20%. Basic amino acids such as Arginine bind preferably at the S1 site, the S2 site sits deeper in the enzyme therefore is restricted to bind hydrophobic amino acids with phenylalanine in the P2 position. Dcp is divided into two subdomains (I, and II), which are the two sides of the clam shell-like structure and has a deep inner cavity where a pair of histidine residues bind to the catalytic zinc ion in the active site. Peptidyl-Dipeptidase Dcp is classified like Angiotensin-I converting enzyme (ACE) which is also a carboxypeptidase involved in blood pressure regulation, but due to structural differences and peptidase activity between these two enzymes they had to be examined separately. ACE has endopeptidase activity, whereas Dcp strictly has exopeptidase activity based on its cytoplasmic location and therefore their mechanisms of action are differentiated. Another difference between these enzymes is that the activity of Peptidyl-Dipeptidase Dcp is not enhanced in the presence of chloride anions, whereas chloride enhances ACE activity.

Scytalidocarboxyl peptidase B, also known as Scytalidoglutamic peptidase and Scytalidopepsin B is a proteolytic enzyme. It was previously thought to be an aspartic protease, but determination of its molecular structure showed it to belong a novel group of proteases, glutamic protease.
Streptomyces cacaoi is a bacterium species from the genus of Streptomyces. Streptomyces cacaoi produces polyoxine.
Streptomyces violaceorectus is a bacterium species from the genus of Streptomyces which has been isolated from soil. Streptomyces violaceorectus produces antimicrobial substances and alkinonase A.
References
- ↑ Kurisu G, Sugimoto A, Harada S, Takagi M, Imanaka T, Kai Y (1997). "Characterization of a small metalloprotease from Streptomyces caespitosus with high specificity to aromatic residues". J. Ferment. Bioeng. 83: 590–592. doi:10.1016/s0922-338x(97)81142-7.
- ↑ Butler MJ, Rawlings ND, Woessner JF (1998). "Snapalysin". In Barrett AJ (ed.). Handbook of Proteolytic Enzymes. London: Handbook of Proteolytic Enzymes. pp. 1134–1135.
- ↑ Kurisu G, Kai Y, Harada S (November 2000). "Structure of the zinc-binding site in the crystal structure of a zinc endoprotease from Streptomyces caespitosus at 1 A resolution". Journal of Inorganic Biochemistry. 82 (1–4): 225–8. doi:10.1016/s0162-0134(00)00136-7. PMID 11132632.