
Gram-negative bacteria are bacteria that do not retain the crystal violet stain used in the Gram staining method of bacterial differentiation. They are characterized by their cell envelopes, which are composed of a thin peptidoglycan cell wall sandwiched between an inner cytoplasmic cell membrane and a bacterial outer membrane.

A spirochaete or spirochete is a member of the phylum Spirochaetes, which contains distinctive diderm (double-membrane) gram-negative bacteria, most of which have long, helically coiled cells. Spirochaetes are chemoheterotrophic in nature, with lengths between 3 and 500 μm and diameters around 0.09 to at least 3 μm.
The Aquificae phylum is a diverse collection of bacteria that live in harsh environmental settings. The name 'Aquificae' was given to this phylum based on an early genus identified within this group, Aquifex, which is able to produce water by oxidizing hydrogen. They have been found in springs, pools, and oceans. They are autotrophs, and are the primary carbon fixers in their environments. These bacteria are Gram-negative, non-spore-forming rods. They are true bacteria as opposed to the other inhabitants of extreme environments, the Archaea.
The Chloroflexia are one of six classes of bacteria in the phylum Chloroflexi, known as filamentous green non-sulfur bacteria. They use light for energy and are named for their green pigment, usually found in photosynthetic bodies called chlorosomes.

Deinococcus–Thermus is a phylum of bacteria with a single order, Deinococci, that are highly resistant to environmental hazards, also known as extremophiles. These bacteria have thick cell walls that give them gram-positive stains, but they include a second membrane and so are closer in structure to those of gram-negative bacteria. Cavalier-Smith calls this clade Hadobacteria.
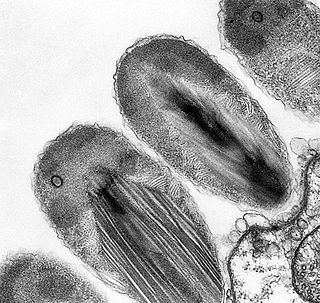
Verrucomicrobia is a phylum of Gram-negative bacteria that contains only a few described species. The species identified have been isolated from fresh water, marine and soil environments and human faeces. A number of as-yet uncultivated species have been identified in association with eukaryotic hosts including extrusive explosive ectosymbionts of protists and endosymbionts of nematodes residing in their gametes.

The Chlamydiae are a bacterial phylum and class whose members are remarkably diverse, including pathogens of humans and animals, symbionts of ubiquitous protozoa, and marine sediment forms not yet well understood. All of the Chlamydiae that humans have known about for many decades are obligate intracellular bacteria; in 2020 many additional Chlamydiae were discovered in ocean-floor environments, and it is not yet known whether they all have hosts. Historically it was believed that all Chlamydiae had a peptidoglycan-free cell wall, but studies in the 2010s demonstrated a detectable presence of peptidoglycan, as well as other important proteins.
The Deferribacteraceae are a family of gram-negative bacteria which make energy by anaerobic respiration.
The Thermotogae are a phylum of the domain Bacteria. The phylum Thermotogae is composed of Gram-negative staining, anaerobic, and mostly thermophilic and hyperthermophilic bacteria.
Fibrobacteres is a small bacterial phylum which includes many of the major rumen bacteria, allowing for the degradation of plant-based cellulose in ruminant animals. Members of this phylum were categorized in other phyla. The genus Fibrobacter was removed from the genus Bacteroides in 1988.
The Veillonellaceae are a family of the Clostridia, formerly known as Acidaminococcaceae. Bacteria in this family are grouped together mainly based on genetic studies, which place them among the Firmicutes. Supporting this placement, several species are capable of forming endospores. However, they differ from most other Firmicutes in having Gram-negative stains. The cell wall composition is peculiar.

Deinococcus is in the monotypic family Deinococcaceae, and one genus of three in the order Deinococcales of the bacterial phylum Deinococcus-Thermus highly resistant to environmental hazards. These bacteria have thick cell walls that give them Gram-positive stains, but they include a second membrane and so are closer in structure to Gram-negative bacteria. Deinococcus survive when their DNA is exposed to high doses of gamma and UV radiation. Whereas other bacteria change their structure in the presence of radiation, such as by forming endospores, Deinococcus tolerate it without changing their cellular form and do not retreat into a hardened structure. They are also characterized by the presence of the carotenoid pigment deinoxanthin that give them their pink color. They are usually isolated according to these two criteria. In August 2020, scientists reported that bacteria from Earth, particularly Deinococcus bacteria, were found to survive for three years in outer space, based on studies conducted on the International Space Station. These findings support the notion of panspermia, the hypothesis that life exists throughout the Universe, distributed in various ways, including space dust, meteoroids, asteroids, comets, planetoids or contaminated spacecraft.
Lentisphaerae is a phylum of bacteria closely related to Chlamydiae and Verrucomicrobia.
The Negativicutes are a class of bacteria in the phylum Firmicutes, whose members have a peculiar cell wall with a lipopolysaccharide outer membrane which stains gram-negative, unlike most other members of the Firmicutes. Although several neighbouring Clostridia species also stain gram-negative, the proteins responsible for the unusual diderm structure of the Negativicutes may have actually been laterally acquired from Proteobacteria. Additional research is required to confirm the origin of the diderm cell envelope in the Negativicutes.
Acidaminococcus is a genus in the phylum Firmicutes (Bacteria), whose members are anaerobic diplococci that can use amino acids as the sole energy source for growth. Like other members of the class Negativicutes, they are gram-negative, despite being Firmicutes, which are normally gram-positive.
The Selenomonadales are an order of bacteria within the class Negativicutes; unlike most other members of Firmicutes, they are Gram-negative. The phylogeny of this order was initially determined by 16S rRNA comparisons. More recently, molecular markers in the form of conserved signature indels (CSIs) have been found specific for all Selenomonadales species. On the basis of these markers, the Selenomonadales are inclusive of two distinct families, and are no longer the sole order within the Negativicutes. Several CSIs have also been found specific for both families, Sporomusaceae and Selenomonadceae. Samples of bacterial strains within this order have been isolated from the root canals of healthy human teeth.
The Coriobacteriia are a class of Gram-positive bacteria within the Actinobacteria phylum. Species within this group are nonsporulating, strict or facultative anaerobes that are capable of thriving in a diverse set of ecological niches. Gordonibacter species are the only members capable of motility by means of flagella within the class. Several species within the Coriobacteriia class have been implicated with human diseases that range in severity. Atopobium, Olsenella, and Cryptobacterium species have responsible for human oral infections including periodontitis, halitosis, and other endodontic infections. Eggerthella species have been associated with severe blood bacteraemia and ulcerative colitis.
Negativicoccus is a Gram-negative and anaerobic genus of bacteria from the family of Veillonellaceae.
Jonquetella is a Gram-negative and strictly aerobic genus of bacteria from the family of Synergistaceae with one known species. Jonquetella anthropi has been isolated from a human cyst from Montpellier in France.
The Eggerthellaceae are a family of Gram-positive, rod- or coccus-shaped Actinobacteria. It is the sole family within the order Eggerthellales.




