Related Research Articles
The Aquificae phylum is a diverse collection of bacteria that live in harsh environmental settings. The name 'Aquificae' was given to this phylum based on an early genus identified within this group, Aquifex, which is able to produce water by oxidizing hydrogen. They have been found in springs, pools, and oceans. They are autotrophs, and are the primary carbon fixers in their environments. These bacteria are Gram-negative, non-spore-forming rods. They are true bacteria as opposed to the other inhabitants of extreme environments, the Archaea.

Deinococcus–Thermus is a phylum of bacteria with a single order, Deinococci, that are highly resistant to environmental hazards, also known as extremophiles. These bacteria have thick cell walls that give them gram-positive stains, but they include a second membrane and so are closer in structure to those of gram-negative bacteria. Cavalier-Smith calls this clade Hadobacteria.

Acidobacteria is a phylum of bacteria. Its members are physiologically diverse and ubiquitous, especially in soils, but are under-represented in culture.
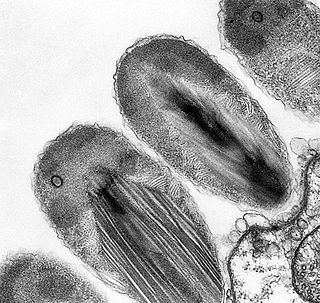
Verrucomicrobia is a phylum of Gram-negative bacteria that contains only a few described species. The species identified have been isolated from fresh water, marine and soil environments and human faeces. A number of as-yet uncultivated species have been identified in association with eukaryotic hosts including extrusive explosive ectosymbionts of protists and endosymbionts of nematodes residing in their gametes.
The Thermotogae are a phylum of the domain Bacteria. The phylum Thermotogae is composed of Gram-negative staining, anaerobic, and mostly thermophilic and hyperthermophilic bacteria.
The Synergistetes is a recently recognized phylum of anaerobic bacteria that show Gram-negative staining and have rod/vibrioid cell shape. Although Synergistetes have a diderm cell envelope, the genes for various proteins involved in lipopolysaccharides biosynthesis have not yet been detected in Synergistetes, indicating that they may have an atypical outer cell envelope. The Synergistetes inhabit a majority of anaerobic environments including animal gastrointestinal tracts, soil, oil wells, and wastewater treatment plants and they are also present in sites of human diseases such as cysts, abscesses, and areas of periodontal disease. Due to their presence at illness related sites, the Synergistetes are suggested to be opportunistic pathogens but they can also be found in healthy individuals in the microbiome of the umbilicus and in normal vaginal flora. Species within this phylum have also been implicated in periodontal disease, gastrointestinal infections and soft tissue infections. Other species from this phylum have been identified as significant contributors in the degradation of sludge for production of biogas in anaerobic digesters and are potential candidates for use in renewable energy production through their production of hydrogen gas. All of the known Synergistetes species and genera are presently part of a single class (Synergistia), order (Synergistiales) and family (Synergistaceae).

Deinococcus is in the monotypic family Deinococcaceae, and one genus of three in the order Deinococcales of the bacterial phylum Deinococcus-Thermus highly resistant to environmental hazards. These bacteria have thick cell walls that give them Gram-positive stains, but they include a second membrane and so are closer in structure to Gram-negative bacteria. Deinococcus survive when their DNA is exposed to high doses of gamma and UV radiation. Whereas other bacteria change their structure in the presence of radiation, such as by forming endospores, Deinococcus tolerate it without changing their cellular form and do not retreat into a hardened structure. They are also characterized by the presence of the carotenoid pigment deinoxanthin that give them their pink color. They are usually isolated according to these two criteria. In August 2020, scientists reported that bacteria from Earth, particularly Deinococcus bacteria, were found to survive for three years in outer space, based on studies conducted on the International Space Station. These findings support the notion of panspermia, the hypothesis that life exists throughout the Universe, distributed in various ways, including space dust, meteoroids, asteroids, comets, planetoids or contaminated spacecraft.
Lentisphaerae is a phylum of bacteria closely related to Chlamydiae and Verrucomicrobia.

Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.
The phylum Elusimicrobia, previously known as "Termite Group 1", has been shown to be widespread in different ecosystems like marine environment, sewage sludge, contaminated sites and soils, and toxic wastes. The high abundance of 'Elusimicrobia' representatives is only evidenced for the lineage of symbionts found in termites and ants.
The Negativicutes are a class of bacteria in the phylum Firmicutes, whose members have a peculiar cell wall with a lipopolysaccharide outer membrane which stains gram-negative, unlike most other members of the Firmicutes. Although several neighbouring Clostridia species also stain gram-negative, the proteins responsible for the unusual diderm structure of the Negativicutes may have actually been laterally acquired from Proteobacteria. Additional research is required to confirm the origin of the diderm cell envelope in the Negativicutes.
Actinocorallia is a genus in the phylum Actinobacteria (Bacteria).
Dactylosporangium is a genus in the phylum Actinobacteria (Bacteria).
Agromyces is a genus in the phylum Actinobacteria (Bacteria).

Gemmatimonas aurantiaca is a Gram-negative, aerobic, polyphosphate-accumulating micro-organism. It is a Gram-negative, rod-shaped aerobe, with type strain T-27T. It replicates by budding.
Chthonomonas calidirosea is a Gram-negative bacterium and also the first representative of the new class Chthonomonadetes within the phylum Armatimonadetes. The Armatimonadetes were previously known as candidate phylum OP10. OP10 was composed solely of environmental 16S rRNA gene clone sequences prior to C. calidirosea's relative, Armatimonas rosea's discovery. It is now known that bacterial communities from geothermal environments, are generally constituted by, at least 5–10% of bacteria belonging to Armatimonadetes.
The Coriobacteriia are a class of Gram-positive bacteria within the Actinobacteria phylum. Species within this group are nonsporulating, strict or facultative anaerobes that are capable of thriving in a diverse set of ecological niches. Gordonibacter species are the only members capable of motility by means of flagella within the class. Several species within the Coriobacteriia class have been implicated with human diseases that range in severity. Atopobium, Olsenella, and Cryptobacterium species have responsible for human oral infections including periodontitis, halitosis, and other endodontic infections. Eggerthella species have been associated with severe blood bacteraemia and ulcerative colitis.
Dyadobacter is a genus of gram negative rod-shaped bacteria belonging to the family Spirosomaceae in the phylum Bacteroidetes. Typical traits of the genus include yellow colony colour, positive flexirubin test and non-motile behaviours. They possess an anaerobic metabolism, can utilise a broad range of carbon sources, and test positive for peroxide catalase activity. The type species is Dyadobacter fermentans, which was isolated from surface sterilised maize leaves,.
Mameliella is a genus in the phylum Proteobacteria (Bacteria). The name Mameliella derives from: New Latin feminine gender dim. noun Mameliella, arbitrary name derived from the acronym MMEL, marine microbial ecology laboratory.
Chitinophagaceae is an aerobic or facultatively anaerobic and rod-shaped family of bacteria in the phylum "Bacteroidetes".
References
- ↑ Zhang H, Sekiguchi Y, Hanada S, Hugenholtz P, Kim H, Kamagata Y, Nakamura K (2003). "Gemmatimonas aurantiaca gen. nov., sp. nov., a gram-negative, aerobic, polyphosphate-accumulating micro-organism, the first cultured representative of the new bacterial phylum Gemmatimonadetes phyl. nov". Int J Syst Evol Microbiol. 53 (Pt 4): 1155–63. doi: 10.1099/ijs.0.02520-0 . PMID 12892144.
- ↑ Takaichi, S; Maoka, T; Takasaki, K; Hanada, S (2009). "Carotenoids of Gemmatimonas aurantiaca (Gemmatimonadetes): identification of a novel carotenoid, deoxyoscillol 2-rhamnoside, and proposed biosynthetic pathway of oscillol 2,2′-dirhamnoside". Microbiology. 156 (3): 757–763. doi: 10.1099/mic.0.034249-0 . PMID 19959572.
- ↑ DeBruyn J.M.; Fawaz M.N.; Peacock, A.D.; Dunlap J.R.; Nixon L.T.; Cooper K.E.; Radosevich M. (2013). "Gemmatirosa kalamazoonesis gen. nov., sp. nov., a member of the rarelycultivated bacterial phylum Gemmatimonadetes". J Gen Appl Microbiol. 59 (4): 305–312. doi: 10.2323/jgam.59.305 . PMID 24005180.
- ↑ Zeng Y.; Selyanin V.; Lukeš M.; Dean J.; Kaftan D.; Feng F.; Koblížek M. (2015). "Characterization of the microaerophilic, bacteriochlorophyll a-containing bacterium Gemmatimonas phototrophica sp. nov., and emended descriptions of the genus Gemmatimonas and Gemmatimonas aurantiaca". Int J Syst Evol Microbiol. 65 (8): 2410–2419. doi: 10.1099/ijs.0.000272 . PMID 25899503.
- ↑ Zeng Y.; Feng F.; Medová H.; Dean J.; Koblížek M. (2014). "Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes". Proc Natl Acad Sci USA. 111 (21): 7795–7800. Bibcode:2014PNAS..111.7795Z. doi: 10.1073/pnas.1400295111 . PMC 4040607 . PMID 24821787.
- ↑ Pascual J.; García-López M.; Bills G.F.; Genilloud O. (2016). "Longimicrobium terrae gen. nov., sp. nov., a novel oligotrophic bacterium of the underrepresented phylum Gemmatimonadetes isolated through a system of miniaturized diffusion chambers". Int J Syst Evol Microbiol. 66 (5): 1976–1985. doi: 10.1099/ijsem.0.000974 . PMID 26873585.
- ↑ Fawaz, Mariam (2013). "Revealing the Ecological Role of Gemmatimonadetes Through Cultivation and Molecular Analysis of Agricultural Soils". Master's Thesis, University of Tennessee: vi.
- ↑ DeBruyn, J; Nixon, L; Fawaz, M; Johnson, M; Radosevich, M (2011). "Global Biogeography and Quantitative Season Dynamics of Gemmatimonadetes in Soil". Appl. Environ. Microbiol. 77 (17): 6295–300. doi:10.1128/AEM.05005-11. PMC 3165389 . PMID 21764958.
- ↑ Fawaz, Mariam (2013). "Revealing the Ecological Role of Gemmatimonadetes Through Cultivation and Molecular Analysis of Agricultural Soils". Master's Thesis, University of Tennessee: vi.
- ↑ J.P. Euzéby. "Gemmatimonadetes". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved 2016-03-20.
- ↑ Sayers; et al. "Gemmatimonadetes". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 2016-03-20.
- ↑ All-Species Living Tree Project "16S rRNA-based LTP release 132". Silva Comprehensive Ribosomal RNA Database . Retrieved 2015-08-20.
- ↑ "GTDB release 05-RS95". Genome Taxonomy Database .
- ↑ Parks, DH; Chuvochina, M; Chaumeil, PA; Rinke, C; Mussig, AJ; Hugenholtz, P (September 2020). "A complete domain-to-species taxonomy for Bacteria and Archaea". Nature Biotechnology. 38 (9): 1079–1086. bioRxiv 10.1101/771964 . doi:10.1038/s41587-020-0501-8. PMID 32341564. S2CID 216560589.