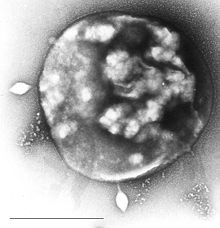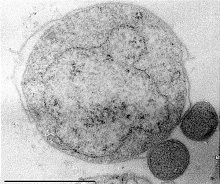
Nanoarchaeota are a phylum of the Archaea. This phylum currently has only one representative, Nanoarchaeum equitans.

In taxonomy, the Korarchaeota are a phylum of the Archaea. The name is derived from the Greek noun koros or kore, meaning young man or young woman, and the Greek adjective archaios which means ancient. They are also known as Xenarchaeota.
The Thermoprotei is a class of the Crenarchaeota.
Mollicutes is a class of bacteria distinguished by the absence of a cell wall. The word "Mollicutes" is derived from the Latin mollis, and cutis. Individuals are very small, typically only 0.2–0.3 μm in size and have a very small genome size. They vary in form, although most have sterols that make the cell membrane somewhat more rigid. Many are able to move about through gliding, but members of the genus Spiroplasma are helical and move by twisting. The best-known genus in the Mollicutes is Mycoplasma.
In taxonomy, the Halobacteriaceae are a family of the Halobacteriales in the domain Archaea. Halobacteriaceae represent a large part of halophilic Archaea, along with members in two other methanogenic families, Methanosarcinaceae and Methanocalculaceae. The family consists of many diverse genera that can survive extreme environmental niches. Most commonly, Halobacteriaceae are found in hypersaline lakes and can even tolerate sites polluted by heavy metals. They include neutrophiles, acidophiles, alkaliphiles, and there have even been psychrotolerant species discovered. Some members have been known to live aerobically, as well as anaerobically, and they come in many different morphologies. These diverse morphologies include rods in genus Halobacterium, cocci in Halococcus, flattened discs or cups in Haloferax, and other shapes ranging from flattened triangles in Haloarcula to squares in Haloquadratum, and Natronorubrum. Most species of Halobacteriaceae are best known for their high salt tolerance and red-pink pigmented members, but there are also non-pigmented species and those that require moderate salt conditions. Some species of Halobacteriaceae have been shown to exhibit phosphorus solubilizing activities that contribute to phosphorus cycling in hypersaline environments. Techniques such as 16S rRNA analysis and DNA-DNA hybridization have been major contributors to taxonomic classification in Halobacteriaceae, partly due to the difficulty in culturing halophilic Archaea.
In taxonomy, the Thermoplasmata are a class of the Euryarchaeota.
In taxonomy, Thermoproteus is a genus of the Thermoproteaceae. These prokaryotes are thermophilic sulphur-dependent organisms related to the genera Sulfolobus, Pyrodictium and Desulfurococcus. They are hydrogen-sulphur autotrophs and can grow at temperatures of up to 95 °C.
Methanococcus is a genus of coccoid methanogens of the family Methanococcaceae. They are all mesophiles, except the thermophilic M. thermolithotrophicus and the hyperthermophilic M. jannaschii. The latter was discovered at the base of a “white smoker” chimney at 21°N on the East Pacific Rise and it was the first archaeal genome to be completely sequenced, revealing many novel and eukaryote-like elements.

In the taxonomy of microorganisms, the Methanomicrobia are a class of the Euryarchaeota.
In the taxonomy of microorganisms, the Methanomicrobiales are an order of the Methanomicrobia. Methanomicrobiales are strictly carbon dioxide reducing methanogens, using hydrogen or formate as the reducing agent. As seen from the phylogenetic tree based on 'The All-Species Living Tree' Project the family Methanomicrobiaceae is highly polyphyletic within the Methanomicrobiales.

In taxonomy, the Sulfolobales are an order of the Thermoprotei.

In taxonomy, the Thermococcales are an order of microbes within the Thermococci. The species within the Thermococcales are used in laboratories as model organisms. All these species are strict anaerobes and can ferment sugars as sources of carbon, but they also need elemental sulfur.
In taxonomy, the Thermoproteales are an order of the Thermoprotei. They are the only organisms known to lack the SSB proteins, instead possessing the protein ThermoDBP that has displaced them. The rRNA genes of these organisms contain multiple introns, which can be homing endonuclease encoding genes, and their presence can impact the binding of "universal" 16S rRNA primers often used in environmental sequencing surveys.
In taxonomy, the Methanocorpusculaceae are a family of microbes within the order Methanomicrobiales. It contains exactly one genus, Methanocorpusculum. The species within Methanocorpusculum were first isolated from anaerobic digesters and anaerobic wastewater treatment plants. In the wild, they prefer freshwater environments. Unlike many other methanogenic archaea, they do not require high temperatures or extreme salt concentrations to live and grow.
In taxonomy, the Methanomicrobiaceae are a family of the Methanomicrobiales.

In taxonomy, the Methanosarcinaceae are a family of the Methanosarcinales.
Methanospirillaceae are a family of microbes within Methanomicrobiales.
Pseudomonas avellanae is a Gram-negative plant pathogenic bacterium. It is the causal agent of bacterial canker of hazelnut. Based on 16S rRNA analysis, P. avellanae has been placed in the P. syringae group. This species was once included as a pathovar of Pseudomonas syringae, but following DNA-DNA hybridization, it was instated as a separate species. Following ribotypical analysis Pseudomonas syringae pv. theae was incorporated into this species.

George Edward Fox is an astrobiologist, a Professor Emeritus and researcher at the University of Houston. He is an elected fellow of the American Academy of Microbiology, the American Association for the Advancement of Science, American Institute for Medical and Biological Engineering and the International Astrobiology Society. Fox received his B.A. degree in 1967, and completed his Ph.D. degree in 1974; both in chemical engineering at Syracuse University.
In taxonomy, Methanocalculus is a genus of the Methanomicrobiales. It contains four species: