- Trypanosoma brucei (Euglenozoa: Kinetoplastida)
- Bodo sp. (Euglenozoa: Kinetoplastida)
- Percolomonas sp. (Percolozoa)
- Stephanopogon sp. (Percolozoa)
- Acrasis rosea (Percolozoa: Heterolobosea)
- Jakobids (Jakobida)
- Giardia sp. (Metamonada: Fornicata: Diplomonadida)
| Excavates Temporal range: | |
|---|---|
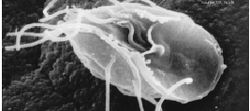 | |
| Giardia lamblia , a parasitic diplomonad | |
| Scientific classification (obsolete as paraphyletic) | |
| Domain: | Eukaryota |
| Infrakingdom: | Excavata (Cavalier-Smith), 2002 |
| Groups included | |
See text | |
| Cladistically included but traditionally excluded taxa | |
All of other Eukaryota | |

Excavata is an obsolete, extensive and diverse paraphyletic group of unicellular Eukaryota. [1] [2] The group was first suggested by Simpson and Patterson in 1999 [3] [4] and the name latinized and assigned a rank by Thomas Cavalier-Smith in 2002. It contains a variety of free-living and symbiotic protists, and includes some important parasites of humans such as Giardia and Trichomonas . [5] Excavates were formerly considered to be included in the now- obsolete Protista kingdom. [6] They were distinguished from other lineages based on electron-microscopic information about how the cells are arranged (they have a distinctive ultrastructural identity). [4] They are considered to be a basal flagellate lineage. [7]
Contents
- Characteristics
- Proposed group
- Discobids or JEH clade
- Metamonads
- Malawimonads
- Ancyromonads
- Evolution
- Origin of the eukaryotes
- Phylogeny
- See also
- Gallery
- References
- External links
On the basis of phylogenomic analyses, the group was shown to contain three widely separated eukaryote groups, the discobids, metamonads, and malawimonads. [8] [9] [10] [11] A current view of the composition of the excavates is given below, indicating that the group is paraphyletic. Except for some Euglenozoa, all are non-photosynthetic.