Related Research Articles
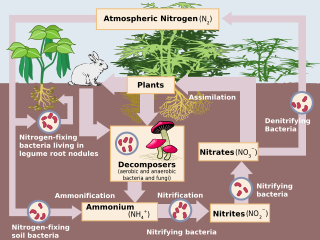
Nitrification is the biological oxidation of ammonia to nitrite followed by the oxidation of the nitrite to nitrate occurring through separate organisms or direct ammonia oxidation to nitrate in comammox bacteria. The transformation of ammonia to nitrite is usually the rate limiting step of nitrification. Nitrification is an important step in the nitrogen cycle in soil. Nitrification is an aerobic process performed by small groups of autotrophic bacteria and archaea.
The Thermomicrobia is a group of thermophilic green non-sulfur bacteria. Based on species Thermomicrobium roseum and Sphaerobacter thermophilus, this bacteria class has the following description:

Planctomycetes are a phylum of widely distributed bacteria, occurring in both aquatic and terrestrial habitats. They play a considerable role in global carbon and nitrogen cycles, with many species of this phylum capable of anaerobic ammonium oxidation, also known as anammox. Many planctomycetes occur in relatively high abundance as biofilms, often associating with other organisms such as macroalgae and marine sponges.

Anammox, an abbreviation for anaerobic ammonium oxidation, is a globally important microbial process of the nitrogen cycle that takes place in many natural environments. The bacteria mediating this process were identified in 1999, and were a great surprise for the scientific community. In the anammox reaction, nitrite and ammonium ions are converted directly into diatomic nitrogen and water.
Methanotrophs are prokaryotes that metabolize methane as their source of carbon and energy. They can be either bacteria or archaea and can grow aerobically or anaerobically, and require single-carbon compounds to survive.
Nitrobacter is a genus comprising rod-shaped, gram-negative, and chemoautotrophic bacteria. The name Nitrobacter derives from the Latin neuter gender noun nitrum, nitri, alkalis; the Ancient Greek noun βακτηρία, βακτηρίᾱς, rod. They are non-motile and reproduce via budding or binary fission. Nitrobacter cells are obligate aerobes and have a doubling time of about 13 hours.
Nitrifying bacteria are chemolithotrophic organisms that include species of the genera e.g. Nitrosomonas, Nitrosococcus, Nitrobacter, Nitrospina, Nitrospira and Nitrococcus. These bacteria get their energy by the oxidation of inorganic nitrogen compounds. Types include ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria (NOB). Many species of nitrifying bacteria have complex internal membrane systems that are the location for key enzymes in nitrification: ammonia monooxygenase, hydroxylamine oxidoreductase, and nitrite oxidoreductase.
Nitrospira translate into “a nitrate spiral” is a genus of bacteria within the monophyletic clade of Nitrospirae phylum. The first member of this genus was described 1986 by Watson et al. isolated from the Gulf of Maine. The bacterium was named Nitrospira marina. Populations were initially thought to be limited to marine ecosystems, but it was later discovered to be well-suited for numerous habitats, including activated sludge of wastewater treatment systems, natural biological marine settings, water circulation biofilters in aquarium tanks, terrestrial systems, fresh and salt water ecosystems, and hot springs. Nitrospira is a ubiquitous bacterium that plays a role in the nitrogen cycle by performing nitrite oxidation in the second step of nitrification. Nitrospira live in a wide array of environments including but not limited to, drinking water systems, waste treatment plants, rice paddies, forest soils, geothermal springs, and sponge tissue. Despite being abundant in many natural and engineered ecosystems Nitrospira are difficult to culture, so most knowledge of them is from molecular and genomic data. However, due to their difficulty to be cultivated in laboratory settings, the entire genome was only sequenced in one species, Nitrospira defluvii. In addition, Nitrospira bacteria's 16s rRNA sequences are too dissimilar to use for PCR primers, thus some members go unnoticed. In addition, members of Nitrospira with the capabilities to perform complete nitrification has also been discovered and cultivated.
The Chloroflexi or Chlorobacteria are a phylum of bacteria containing isolates with a diversity of phenotypes, including members that are aerobic thermophiles, which use oxygen and grow well in high temperatures; anoxygenic phototrophs, which use light for photosynthesis ; and anaerobic halorespirers, which uses halogenated organics as electron acceptors.
The Thaumarchaeota or Thaumarchaea are a phylum of the Archaea proposed in 2008 after the genome of Cenarchaeum symbiosum was sequenced and found to differ significantly from other members of the hyperthermophilic phylum Crenarchaeota. Three described species in addition to C. symbosium are Nitrosopumilus maritimus, Nitrososphaera viennensis, and Nitrososphaera gargensis. The phylum was proposed in 2008 based on phylogenetic data, such as the sequences of these organisms' ribosomal RNA genes, and the presence of a form of type I topoisomerase that was previously thought to be unique to the eukaryotes. This assignment was confirmed by further analysis published in 2010 that examined the genomes of the ammonia-oxidizing archaea Nitrosopumilus maritimus and Nitrososphaera gargensis, concluding that these species form a distinct lineage that includes Cenarchaeum symbiosum. The lipid crenarchaeol has been found only in Thaumarchaea, making it a potential biomarker for the phylum. Most organisms of this lineage thus far identified are chemolithoautotrophic ammonia-oxidizers and may play important roles in biogeochemical cycles, such as the nitrogen cycle and the carbon cycle. Metagenomic sequencing indicates that they constitute ~1% of the sea surface metagenome across many sites.

Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.
"Candidatus Scalindua" is a bacterial genus, and a proposed member of the order Planctomycetes. These bacteria lack peptidoglycan in their cell wall and have a compartmentalized cytoplasm. They are ammonium oxidizing bacteria found in marine environments.
Nitrospira moscoviensis was the second bacterium classified under the most diverse nitrite-oxidizing bacteria phylum, Nitrospirae. It is a gram-negative, non-motile, facultative lithoauthotropic bacterium that was discovered in Moscow, Russia in 1995. The genus name, Nitrospira, originates from the prefix “nitro” derived from nitrite, the microbe’s electron donor and “spira” meaning coil or spiral derived from the microbe’s shape. The species name, moscoviensis, is derived from Moscow, where the species was first discovered. N. moscoviensis could potentially be used in the production of bio-degradable polymers.
Comammox is the name attributed to an organism that can convert ammonia into nitrite and then into nitrate through the process of nitrification. Nitrification has traditionally thought to be a two-step process, where ammonia-oxidizing bacteria and archaea oxidize ammonia to nitrite and then nitrite-oxidizing bacteria convert to nitrate. Complete conversion of ammonia into nitrate by a single microorganism was first predicted in 2006. In 2015 the presence of microorganisms that could carry out both conversion processes was discovered within the genus Nitrospira, and the nitrogen cycle was updated. Within the genus Nitrospira, the major ecosystems comammox are primarily found in natural aquifers and engineered ecosystems.
Nitrososphaera gargensis is a non-pathogenic, small coccus measuring 0.9 ± 0.3 μm in diameter. N. gargensis is observed in small abnormal cocci groupings and uses its archaella to move via chemotaxis. Being an Archaeon, Nitrososphaera gargensis has a cell membrane composed of crenarchaeol, its isomer, and a distinct glycerol dialkyl glycerol tetraether (GDGT), which is significant in identifying ammonia-oxidizing archaea (AOA). The organism plays a role in influencing ocean communities and food production.

Ubiquitin Bacterial (UBact) is a protein that is homologous to Prokaryotic ubiquitin-like protein (Pup). UBact was recently described by the group of Professor Aaron Ciechanover at the Technion, Israel.
Modulibacteria is a bacterial phylum formerly known as KS3B3 or GN06. It is a candidate phylum, meaning there are no cultured representatives of this group. Members of the Modulibacteria phylum are known to cause fatal filament overgrowth (bulking) in high-rate industrial anaerobic wastewater treatment bioreactors.

NC10 is a bacterial phylum with candidate status, meaning its members remain uncultured to date. The difficulty in producing lab cultures may be linked to low growth rates and other limiting growth factors.
Nitrospira inopinata is a bacterium from the phylum Nitrospirae. This phylum contains nitrite-oxidizing bacteria playing role in nitrification. However N. inopinata was shown to perform complete ammonia oxidation to nitrate thus being the first comammox bacterium to be discovered.
Nitrospinae is a bacterial phylum. Despite only few described species, members of this phylum are major nitrite-oxidizing bacteria in surface waters in oceans. By oxidation of nitrite to nitrate they are important in the process of nitrification in marine environments.
References
- ↑ Watson SW, Bock E, Valois FW, Waterbury JB, Schlosser U (1986). "Nitrospira marina gen. nov. sp. nov.: a chemolithotrophic nitrite-oxidizing bacterium". Arch Microbiol. 144 (1): 1–7. doi:10.1007/BF00454947. S2CID 29796511.
- ↑ Ehrich S, Behrens D, Lebedeva E, Ludwig W, Bock E (July 1995). "A new obligately chemolithoautotrophic, nitrite-oxidizing bacterium, Nitrospira moscoviensis sp. nov. and its phylogenetic relationship". Archives of Microbiology. 164 (1): 16–23. doi:10.1007/BF02568729. PMID 7646315. S2CID 2702110.
- 1 2 Sayers. "Nitrospirae". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 2016-03-20.
- ↑ Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M (November 2001). "In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants". Applied and Environmental Microbiology. 67 (11): 5273–84. doi:10.1128/AEM.67.11.5273-5284.2001. PMC 93301 . PMID 11679356.
- ↑ Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, et al. (December 2015). "Complete nitrification by Nitrospira bacteria". Nature. 528 (7583): 504–9. Bibcode:2015Natur.528..504D. doi:10.1038/nature16461. PMC 5152751 . PMID 26610024.
- ↑ van Kessel MA, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJ, Kartal B, et al. (December 2015). "Complete nitrification by a single microorganism". Nature. 528 (7583): 555–9. Bibcode:2015Natur.528..555V. doi:10.1038/nature16459. PMC 4878690 . PMID 26610025.
- ↑ Kits KD, Sedlacek CJ, Lebedeva EV, Han P, Bulaev A, Pjevac P, et al. (September 2017). "Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle". Nature. 549 (7671): 269–272. Bibcode:2017Natur.549..269K. doi:10.1038/nature23679. PMC 5600814 . PMID 28847001.
- ↑ All-Species Living Tree Project. "16S rRNA-based LTP release 132". Silva Comprehensive Ribosomal RNA Database . Retrieved 2015-08-20.
- ↑ "AnnoTree v1.2.0". AnnoTree.
- ↑ Mendler, K; Chen, H; Parks, DH; Hug, LA; Doxey, AC (2019). "AnnoTree: visualization and exploration of a functionally annotated microbial tree of life". Nucleic Acids Research. 47 (9): 4442–4448. doi: 10.1093/nar/gkz246 . PMC 6511854 . PMID 31081040.
- ↑ "GTDB release 05-RS95". Genome Taxonomy Database .
- ↑ Parks, DH; Chuvochina, M; Chaumeil, PA; Rinke, C; Mussig, AJ; Hugenholtz, P (September 2020). "A complete domain-to-species taxonomy for Bacteria and Archaea". Nature Biotechnology. 38 (9): 1079–1086. bioRxiv 10.1101/771964 . doi:10.1038/s41587-020-0501-8. PMID 32341564. S2CID 216560589.
- ↑ Euzéby JP. ""Nitrospirae"". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved 2016-03-20.