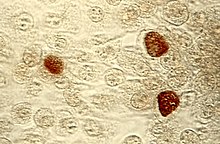
Chlamydia, or more specifically a chlamydia infection, is a sexually transmitted infection caused by the bacterium Chlamydia trachomatis. Most people who are infected have no symptoms. When symptoms do appear they may occur only several weeks after infection; the incubation period between exposure and being able to infect others is thought to be on the order of two to six weeks. Symptoms in women may include vaginal discharge or burning with urination. Symptoms in men may include discharge from the penis, burning with urination, or pain and swelling of one or both testicles. The infection can spread to the upper genital tract in women, causing pelvic inflammatory disease, which may result in future infertility or ectopic pregnancy.
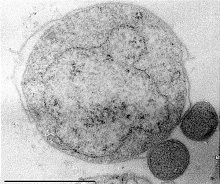
Nanoarchaeota are a phylum of the Archaea. This phylum currently has only one representative, Nanoarchaeum equitans.

Chlamydia trachomatis, commonly known as chlamydia, is a bacterium that causes chlamydia, which can manifest in various ways, including: trachoma, lymphogranuloma venereum, nongonococcal urethritis, cervicitis, salpingitis, pelvic inflammatory disease. C. trachomatis is the most common infectious cause of blindness and the most common sexually transmitted bacterium.
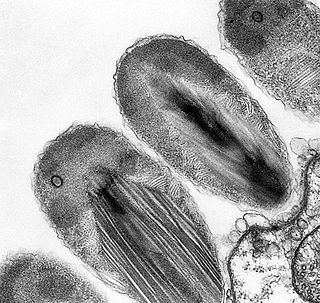
Verrucomicrobia is a phylum of Gram-negative bacteria that contains only a few described species. The species identified have been isolated from fresh water, marine and soil environments and human faeces. A number of as-yet uncultivated species have been identified in association with eukaryotic hosts including extrusive explosive ectosymbionts of protists and endosymbionts of nematodes residing in their gametes.
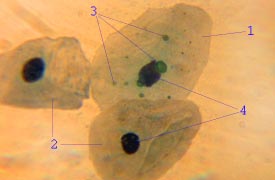
Chlamydia pneumoniae is a species of Chlamydia, an obligate intracellular bacterium that infects humans and is a major cause of pneumonia. It was known as the Taiwan acute respiratory agent (TWAR) from the names of the two original isolates – Taiwan (TW-183) and an acute respiratory isolate designated AR-39. Briefly, it was known as Chlamydophila pneumoniae, and that name is used as an alternate in some sources. In some cases, to avoid confusion, both names are given.

Chlamydia psittaci is a lethal intracellular bacterial species that may cause endemic avian chlamydiosis, epizootic outbreaks in mammals, and respiratory psittacosis in humans. Potential hosts include feral birds and domesticated poultry, as well as cattle, pigs, sheep, and horses. C. psittaci is transmitted by inhalation, contact, or ingestion among birds and to mammals. Psittacosis in birds and in humans often starts with flu-like symptoms and becomes a life-threatening pneumonia. Many strains remain quiescent in birds until activated by stress. Birds are excellent, highly mobile vectors for the distribution of chlamydia infection, because they feed on, and have access to, the detritus of infected animals of all sorts.

Chlamydophila is a controversial bacterial genus belonging to the family Chlamydiaceae, order Chlamydiales, class/phylum Chlamydiae.

The Chlamydiaceae are a family of gram-negative bacteria that belongs to the phylum Chlamydiae, order Chlamydiales. Chlamydiaceae species express the family-specific lipopolysaccharide epitope αKdo-(2→8)-αKdo-(2→4)-αKdo. Chlamydiaceae ribosomal RNA genes all have at least 90% DNA sequence identity. Chlamydiaceae species have varying inclusion morphology, varying extrachromosomal plasmid content, and varying sulfadiazine resistance.
Chlamydia muridarum is an intracellular bacterial species that at one time belonged to Chlamydia trachomatis. However, C. trachomatis naturally only infects humans and C. muridarum naturally infects only members of the family Muridae.
Chlamydia suis is a member of the genus Chlamydia. C. suis has only been isolated from swine, in which it may be endemic. Glycogen has been detected in Chlamydia suis inclusions in infected swine tissues and in cell culture. C. suis is associated with conjunctivitis, enteritis and pneumonia in swine.
Parachlamydiaceae is a family of bacteria in the order Chlamydiales. Species in this family have a Chlamydia–like cycle of replication and their ribosomal RNA genes are 80–90% identical to ribosomal genes in the Chlamydiaceae. The Parachlamydiaceae naturally infect amoebae and can be grown in cultured Vero cells. The Parachlamydiaceae are not recognized by monoclonal antibodies that detect Chlamydiaceae lipopolysaccharide.
Candidatus Rhabdochlamydia is a genus of intracellular bacteria; a separate family, Candidatus Rhabdochlamydiaceae, has been proposed for this genus. Candidatus Rhabdochlamydia species have not been cultured in vitro and have not been deposited in culture collections.

Chlamydia abortus is a species in Chlamydiae that causes abortion and fetal death in mammals, including humans. Chlamydia abortus was renamed in 1999 as Chlamydophila psittaci along with all Chlamydiae except Chlamydia trachomatis. This was based on a lack of evident glycogen production and on resistance to the antibiotic sulfadiazine. In 1999 C. psittaci and C. abortus were recognized as distinct species based on differences of pathogenicity and DNA–DNA hybridization. In 2015, this new name was reverted to Chlamydia.
Chlamydia felis is a Gram-negative, obligate intracellular bacterial pathogen that infects cats. It is endemic among domestic cats worldwide, primarily causing inflammation of feline conjunctiva, rhinitis and respiratory problems. C. felis can be recovered from the stomach and reproductive tract. Zoonotic infection of humans with C. felis has been reported. Strains FP Pring and FP Cello have an extrachromosomal plasmid, whereas the FP Baker strain does not. FP Cello produces lethal disease in mice, whereas the FP Baker does not. An attenuated FP Baker strain, and an attenuated 905 strain, are used as live vaccines for cats.
Chlamydia caviae is a bacterium that can be recovered from the conjunctiva of Guinea pigs suffering from ocular inflammation and eye discharge. It is also possible to infect the genital tract of Guinea pigs with C. caviae and elicit a disease that is very similar to human Chlamydia trachomatis infection. C. caviae infects primarily the mucosal epithelium and is not invasive.
Chlamydia pecorum, also known as Chlamydophila pecorum is a species of Chlamydiaceae that originated from ruminants, such as cattle, sheep and goats. It has also infected koalas and swine. C. pecorum strains are serologically and pathogenically diverse.

Chlamydia is a genus of pathogenic Gram-negative bacteria that are obligate intracellular parasites. Chlamydia infections are the most common bacterial sexually transmitted diseases in humans and are the leading cause of infectious blindness worldwide.
Chlamydia is a sexually transmitted infection caused by the bacterium Chlamydia trachomatis.
Parachlamydia acanthamoebae are bacterium that fall into the category of host-associated microorganisms. This bacterium lives within free-living amoebae that are an intricate part of their reproduction. Originally named Candidatus Parachlamydia acanthamoebae, its current scientific name was introduced shortly after. This species has shown to have over eighty percent 16S rRNA gene sequencing identity with the class Chlamydiae. Parachlamydia acanthamoebae has the same family as the genus Neochlamydia with which it shares many similarities.
Chlamydia research is the systematic study of the organisms in the taxonomic group of bacteria Chlamydiae, the diagnostic procedures to treat infections, the disease chlamydia, infections caused by the organisms, the epidemiology of infection and the development of vaccines. The process of research can include the participation of many researchers who work in collaboration from separate organizations, governmental entities and universities.