In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.
Shikimic acid, more commonly known as its anionic form shikimate, is a cyclohexene, a cyclitol and a cyclohexanecarboxylic acid. It is an important biochemical metabolite in plants and microorganisms. Its name comes from the Japanese flower shikimi, from which it was first isolated in 1885 by Johan Fredrik Eykman. The elucidation of its structure was made nearly 50 years later.
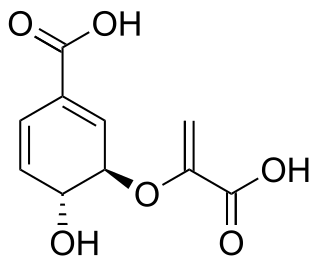
Chorismic acid, more commonly known as its anionic form chorismate, is an important biochemical intermediate in plants and microorganisms. It is a precursor for:

Amino acid synthesis is the set of biochemical processes by which the amino acids are produced. The substrates for these processes are various compounds in the organism's diet or growth media. Not all organisms are able to synthesize all amino acids. For example, humans can synthesize 11 of the 20 standard amino acids. These 11 are called the non-essential amino acids).

Erythrose 4-phosphate is a phosphate of the simple sugar erythrose. It is an intermediate in the pentose phosphate pathway and the Calvin cycle.

In enzymology, a shikimate dehydrogenase (EC 1.1.1.25) is an enzyme that catalyzes the chemical reaction

In enzymology, a phosphoribosylanthranilate isomerase (PRAI) is an enzyme that catalyzes the third step of the synthesis of the amino acid tryptophan.

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction

The enzyme 3-dehydroquinate dehydratase (EC 4.2.1.10) catalyzes the chemical reaction
The enzyme chorismate synthase catalyzes the chemical reaction

Shikimate kinase (EC 2.7.1.71) is an enzyme that catalyzes the ATP-dependent phosphorylation of shikimate to form shikimate 3-phosphate. This reaction is the fifth step of the shikimate pathway, which is used by plants and bacteria to synthesize the common precursor of aromatic amino acids and secondary metabolites. The systematic name of this enzyme class is ATP:shikimate 3-phosphotransferase. Other names in common use include shikimate kinase (phosphorylating), and shikimate kinase II.
The Aminoshikimate pathway is a biochemical pathway present in some plants, which has been studied by biologists, biochemists and especially those interested in manufacture of novel antibiotic drugs. The pathway is a novel variation of the shikimate pathway. The aminoshikimate pathway was first discovered and studied in the rifamycin B producer Amycolatopsis mediterranei. Its end product, 3-amino-5-hydroxybenzoate, serves as an initiator for polyketide synthases in the biosynthesis of ansamycins.
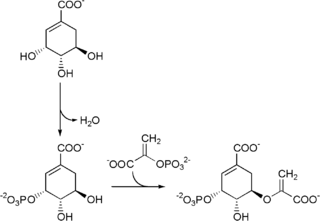
The shikimate pathway is a seven-step metabolic pathway used by bacteria, archaea, fungi, algae, some protozoans, and plants for the biosynthesis of folates and aromatic amino acids. This pathway is not found in mammals.
3-Deoxy-D-arabinoheptulosonate 7-phosphate (DAHP) synthase is the first enzyme in a series of metabolic reactions known as the shikimate pathway, which is responsible for the biosynthesis of the amino acids phenylalanine, tyrosine, and tryptophan. Since it is the first enzyme in the shikimate pathway, it controls the amount of carbon entering the pathway. Enzyme inhibition is the primary method of regulating the amount of carbon entering the pathway. Forms of this enzyme differ between organisms, but can be considered DAHP synthase based upon the reaction that is catalyzed by this enzyme.
5-enolpyruvylshikimate-3-phosphate (EPSP) synthase is an enzyme produced by plants and microorganisms. EPSPS catalyzes the chemical reaction:

3-Deoxy-D-arabino-heptulosonic acid 7-phosphate (DAHP) is a 7-carbon ulosonic acid. This compound is found in the shikimic acid biosynthesis pathway and is an intermediate in the production of aromatic amino acids.
3-dehydroquinate synthase II (EC 1.4.1.24, DHQ synthase II, MJ1249 (gene), aroB' (gene)) is an enzyme with systematic name 2-amino-3,7-dideoxy-D-threo-hept-6-ulosonate:NAD+ oxidoreductase (deaminating). This enzyme catalyses the following chemical reaction
6-deoxy-5-ketofructose 1-phosphate synthase is an enzyme with systematic name 2-oxopropanal:D-fructose 1,6-bisphosphate glycerone-phosphotransferase. This enzyme catalyses the following chemical reaction
2-amino-3,7-dideoxy-D-threo-hept-6-ulosonate synthase is an enzyme with systematic name L-aspartate 4-semialdehyde:1-deoxy-D-threo-hexo-2,5-diulose 6-phosphate methylglyoxaltransferase. This enzyme catalyses the following chemical reaction
2-deoxy-scyllo-Inosose synthase is an enzyme with systematic name D-glucose-6-phosphate phosphate-lyase (2-deoxy-scyllo-inosose-forming). This enzyme catalyses the following chemical reaction



















