Related Research Articles

Angiotensin-converting enzyme, or ACE, is a central component of the renin–angiotensin system (RAS), which controls blood pressure by regulating the volume of fluids in the body. It converts the hormone angiotensin I to the active vasoconstrictor angiotensin II. Therefore, ACE indirectly increases blood pressure by causing blood vessels to constrict. ACE inhibitors are widely used as pharmaceutical drugs for treatment of cardiovascular diseases.

Cathepsin C (CTSC) also known as dipeptidyl peptidase I (DPP-I) is a lysosomal exo-cysteine protease belonging to the peptidase C1 protein family, a subgroup of the cysteine cathepsins. In humans, it is encoded by the CTSC gene.
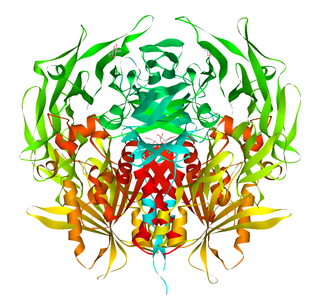
Dipeptidyl peptidase-4 (DPP4), also known as adenosine deaminase complexing protein 2 or CD26 is a protein that, in humans, is encoded by the DPP4 gene. DPP4 is related to FAP, DPP8, and DPP9. The enzyme was discovered in 1966 by Hopsu-Havu and Glenner, and as a result of various studies on chemism, was called dipeptidyl peptidase IV [DP IV].
In molecular biology, the Signal Peptide Peptidase (SPP) is a type of protein that specifically cleaves parts of other proteins. It is an intramembrane aspartyl protease with the conserved active site motifs 'YD' and 'GxGD' in adjacent transmembrane domains (TMDs). Its sequences is highly conserved in different vertebrate species. SPP cleaves remnant signal peptides left behind in membrane by the action of signal peptidase and also plays key roles in immune surveillance and the maturation of certain viral proteins.

Aspartic proteases are a catalytic type of protease enzymes that use an activated water molecule bound to one or more aspartate residues for catalysis of their peptide substrates. In general, they have two highly conserved aspartates in the active site and are optimally active at acidic pH. Nearly all known aspartyl proteases are inhibited by pepstatin.

Thermolysin is a thermostable neutral metalloproteinase enzyme produced by the Gram-positive bacteria Bacillus thermoproteolyticus. It requires one zinc ion for enzyme activity and four calcium ions for structural stability. Thermolysin specifically catalyzes the hydrolysis of peptide bonds containing hydrophobic amino acids. However thermolysin is also widely used for peptide bond formation through the reverse reaction of hydrolysis. Thermolysin is the most stable member of a family of metalloproteinases produced by various Bacillus species. These enzymes are also termed 'neutral' proteinases or thermolysin -like proteinases (TLPs).

N-Acetylaspartylglutamic acid is a peptide neurotransmitter and the third-most-prevalent neurotransmitter in the mammalian nervous system. NAAG consists of N-acetylaspartic acid (NAA) and glutamic acid coupled via a peptide bond.
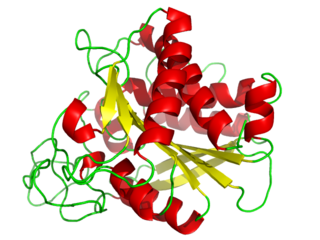
Carboxypeptidase A usually refers to the pancreatic exopeptidase that hydrolyzes peptide bonds of C-terminal residues with aromatic or aliphatic side-chains. Most scientists in the field now refer to this enzyme as CPA1, and to a related pancreatic carboxypeptidase as CPA2.

Xaa-Pro dipeptidase, also known as prolidase, is an enzyme that in humans is encoded by the PEPD gene.

Dipeptidase 1 (DPEP1), or renal dipeptidase, is a membrane-bound glycoprotein responsible for hydrolyzing dipeptides. It is found in the microsomal fraction of the porcine kidney cortex. It exists as a disulfide-linked homodimer that is glygosylphosphatidylinositol (GPI)-anchored to the renal brush border of the kidney. The active site on each homodimer is made up of a barrel subunit with binuclear zinc ions that are bridged by the Gly125 side-chain located at the bottom of the barrel.

Dipeptidyl peptidase I is an enzyme. This enzyme catalyses the following chemical reaction
Dipeptidyl-peptidase III is an enzyme. This enzyme catalyses the following chemical reaction
Xaa-Trp aminopeptidase is an enzyme. This enzyme catalyses the following chemical reaction
PepB aminopeptidase is an enzyme which catalyses the following chemical reaction:

Xaa-Pro dipeptidase is an enzyme. This enzyme catalyses the following chemical reaction

Cytosolic non-specific dipeptidase also known as carnosine dipeptidase 2 is an enzyme that in humans is encoded by the CNDP2 gene. This enzyme catalyses the hydrolysis of dipeptides, preferentially those containing hydrophobic amino acids. The human enzyme uses manganese ions as a cofactor. In addition to its function as a peptidase, the enzyme also functions to generate N-lactoyl amino acids, such as N-lactoyl-phenylalanine, via the process of "reverse proteolysis".
Peptidyl-dipeptidase B is an enzyme. It catalyses the following chemical reaction
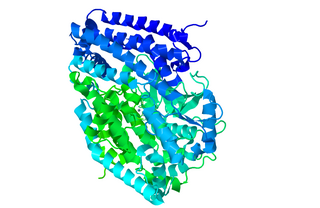
Peptidyl-dipeptidase Dcp (EC 3.4.15.5, dipeptidyl carboxypeptidase (Dcp), dipeptidyl carboxypeptidase) is a metalloenzyme found in the cytoplasm of bacterium E. Coli responsible for the C-terminal cleavage of a variety of dipeptides and unprotected larger peptide chains. The enzyme does not hydrolyze bonds in which P1' is Proline, or both P1 and P1' are Glycine. Dcp consists of 680 amino acid residues that form into a single active monomer which aids in the intracellular degradation of peptides. Dcp coordinates to divalent zinc which sits in the pocket of the active site and is composed of four subsites: S1’, S1, S2, and S3, each subsite attracts certain amino acids at a specific position on the substrate enhancing the selectivity of the enzyme. The four subsites detect and bind different amino acid types on the substrate peptide in the P1 and P2 positions. Some metallic divalent cations such as Ni+2, Cu+2, and Zn+2 inhibit the function of the enzyme around 90%, whereas other cations such as Mn+2, Ca+2, Mg+2, and Co+2 have slight catalyzing properties, and increase the function by around 20%. Basic amino acids such as Arginine bind preferably at the S1 site, the S2 site sits deeper in the enzyme therefore is restricted to bind hydrophobic amino acids with phenylalanine in the P2 position. Dcp is divided into two subdomains (I, and II), which are the two sides of the clam shell-like structure and has a deep inner cavity where a pair of histidine residues bind to the catalytic zinc ion in the active site. Peptidyl-Dipeptidase Dcp is classified like Angiotensin-I converting enzyme (ACE) which is also a carboxypeptidase involved in blood pressure regulation, but due to structural differences and peptidase activity between these two enzymes they had to be examined separately. ACE has endopeptidase activity, whereas Dcp strictly has exopeptidase activity based on its cytoplasmic location and therefore their mechanisms of action are differentiated. Another difference between these enzymes is that the activity of Peptidyl-Dipeptidase Dcp is not enhanced in the presence of chloride anions, whereas chloride enhances ACE activity.
Pyroglutamyl-peptidase I (EC 3.4.19.3, also known as Pyrrolidonyl peptidase, is an enzyme found in bacteria, plants and animals.
Beta-aspartyl-peptidase is an enzyme. This enzyme catalyses the following chemical reaction
References
- ↑ Håkansson K, Wang AH, Miller CG (December 2000). "The structure of aspartyl dipeptidase reveals a unique fold with a Ser-His-Glu catalytic triad". Proceedings of the National Academy of Sciences of the United States of America. 97 (26): 14097–102. Bibcode:2000PNAS...9714097H. doi: 10.1073/pnas.260376797 . PMC 18877 . PMID 11106384.
- ↑ Lassy RA, Miller CG (May 2000). "Peptidase E, a peptidase specific for N-terminal aspartic dipeptides, is a serine hydrolase". Journal of Bacteriology. 182 (9): 2536–43. doi:10.1128/jb.182.9.2536-2543.2000. PMC 111318 . PMID 10762256.