Function
Cathepsin C appears to be a central coordinator for activation of many serine proteases in immune/inflammatory cells.
Cathepsin C catalyses excision of dipeptides from the N-terminus of protein and peptide substrates, except if (i) the amino group of the N-terminus is blocked, (ii) the site of cleavage is on either side of a proline residue, (iii) the N-terminal residue is lysine or arginine, or (iv) the structure of the peptide or protein prevents further digestion from the N-terminus.
Inflammatory response
Particularly, it is involved in activation of neutrophil serine proteases (NSPs; i.e., cathepsin G, proteinase 3 and neutrophil elastase) as they are synthesised as inactive proenzymes during neutrophil maturation. Then, they are released during degranulation. [7] [8] Other enzymes activated by cathepsin C are: chymase and tryptase in mast cells and granzymes A and B, cathepsin G, and elastase in lymphocytes and natural killer cells (NK cells). [9]
Overactivation of NSPs causes a cascade of processess that result in excessive lung inflammation and reduced pathogen clearance. They involve reduced secretion of antileukoproteinase, extracellular matrix degradation, activation of IL-1β, IL-8 and TNF-α as well as inhibition of alpha-1 antitrypsin, an enzyme involved in NSP degradation. [8]
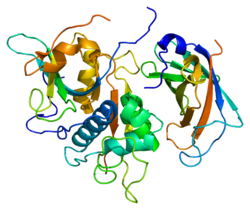
Structure
The cDNAs encoding rat, human, murine, bovine, dog and two Schistosome cathepsin Cs have been cloned and sequenced and show that the enzyme is highly conserved. [10] The human and rat cathepsin C cDNAs encode precursors (prepro-cathepsin C) comprising signal peptides of 24 residues, pro-regions of 205 (rat cathepsin C) or 206 (human cathepsin C) residues and catalytic domains of 233 residues which contain the catalytic residues and are 30–40% identical to the mature amino acid sequences of papain and a number of other cathepsins including cathepsins, B, H, K, L, and S. [11]
The translated prepro-cathepsin C is processed into the mature form by at least four cleavages of the polypeptide chain. The signal peptide is removed during translocation or secretion of the pro-enzyme (pro-cathepsin C) and a large N-terminal proregion fragment (also known as the exclusion domain), [12] which is retained in the mature enzyme, is separated from the catalytic domain by excision of a minor C-terminal part of the pro-region, called the activation peptide. A heavy chain of about 164 residues and a light chain of about 69 residues are generated by cleavage of the catalytic domain.
Unlike the other members of the papain family, mature cathepsin C consists of four subunits, each composed of the N-terminal proregion fragment, the heavy chain and the light chain. Both the pro-region fragment and the heavy chain are glycosylated.
This page is based on this
Wikipedia article Text is available under the
CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.