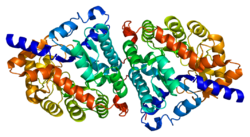
Beta-lactamases, (β-lactamases) are enzymes produced by bacteria that provide multi-resistance to beta-lactam antibiotics such as penicillins, cephalosporins, cephamycins, monobactams and carbapenems (ertapenem), although carbapenems are relatively resistant to beta-lactamase. Beta-lactamase provides antibiotic resistance by breaking the antibiotics' structure. These antibiotics all have a common element in their molecular structure: a four-atom ring known as a beta-lactam (β-lactam) ring. Through hydrolysis, the enzyme lactamase breaks the β-lactam ring open, deactivating the molecule's antibacterial properties.

Angiotensin-converting enzyme, or ACE, is a central component of the renin–angiotensin system (RAS), which controls blood pressure by regulating the volume of fluids in the body. It converts the hormone angiotensin I to the active vasoconstrictor angiotensin II. Therefore, ACE indirectly increases blood pressure by causing blood vessels to constrict. ACE inhibitors are widely used as pharmaceutical drugs for treatment of cardiovascular diseases.

DD-transpeptidase is a bacterial enzyme that catalyzes the transfer of the R-L-αα-D-alanyl moiety of R-L-αα-D-alanyl-D-alanine carbonyl donors to the γ-OH of their active-site serine and from this to a final acceptor. It is involved in bacterial cell wall biosynthesis, namely, the transpeptidation that crosslinks the peptide side chains of peptidoglycan strands.

Piperacillin is a broad-spectrum β-lactam antibiotic of the ureidopenicillin class. The chemical structure of piperacillin and other ureidopenicillins incorporates a polar side chain that enhances penetration into Gram-negative bacteria and reduces susceptibility to cleavage by Gram-negative beta lactamase enzymes. These properties confer activity against the important hospital pathogen Pseudomonas aeruginosa. Thus piperacillin is sometimes referred to as an "anti-pseudomonal penicillin".
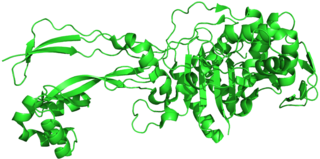
Penicillin-binding proteins (PBPs) are a group of proteins that are characterized by their affinity for and binding of penicillin. They are a normal constituent of many bacteria; the name just reflects the way by which the protein was discovered. All β-lactam antibiotics bind to PBPs, which are essential for bacterial cell wall synthesis. PBPs are members of a subgroup of enzymes called transpeptidases. Specifically, PBPs are DD-transpeptidases.

Leucyl/cystinyl aminopeptidase, also known as cystinyl aminopeptidase (CAP), insulin-regulated aminopeptidase (IRAP), human placental leucine aminopeptidase (PLAP), oxytocinase, and vasopressinase, is an enzyme of the aminopeptidase group that in humans is encoded by the LNPEP gene.

Doripenem is an antibiotic drug in the carbapenem class. It is a beta-lactam antibiotic drug able to kill Pseudomonas aeruginosa.
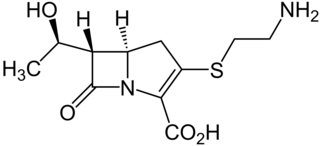
Thienamycin is one of the most potent naturally produced antibiotics known thus far, discovered in Streptomyces cattleya in 1976. Thienamycin has excellent activity against both Gram-positive and Gram-negative bacteria and is resistant to bacterial β-lactamase enzymes. Thienamycin is a zwitterion at pH 7.
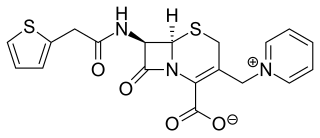
Cephaloridine is a first-generation semisynthetic derivative of antibiotic cephalosporin C. It is a Beta lactam antibiotic, like penicillin. Its chemical structure contains 3 cephems, 4 carboxyl groups and three pyridinium methyl groups.

Gastric lipase, also known as LIPF, is an enzymatic protein that, in humans, is encoded by the LIPF gene.

Xaa-Pro dipeptidase, also known as prolidase, is an enzyme that in humans is encoded by the PEPD gene.

Carnosinemia is a rare autosomal recessive metabolic disorder caused by a deficiency of carnosinase, a dipeptidase.

Dipeptidase 2 (DPEP2) is a protein which in humans is encoded by the DPEP2 gene.

Dipeptidase 3 (DPEP3) is a protein that in humans is encoded by the DPEP3 gene.
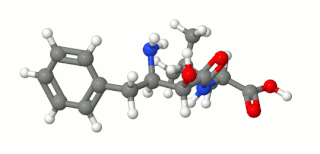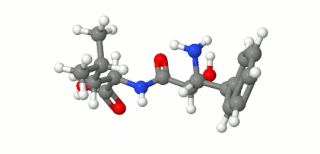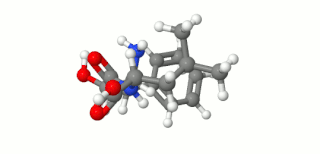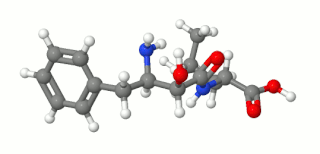
Ubenimex (INN), also known more commonly as bestatin, is a competitive, reversible protease inhibitor. It is an inhibitor of arginyl aminopeptidase (aminopeptidase B), leukotriene A4 hydrolase (a zinc metalloprotease that displays both epoxide hydrolase and aminopeptidase activities), alanyl aminopeptidase (aminopeptidase M/N), leucyl/cystinyl aminopeptidase (oxytocinase/vasopressinase), and membrane dipeptidase (leukotriene D4 hydrolase). It is being studied for use in the treatment of acute myelocytic leukemia and lymphedema. It is derived from Streptomyces olivoreticuli. Ubenimex has been found to inhibit the enzymatic degradation of oxytocin, vasopressin, enkephalins, and various other peptides and compounds.
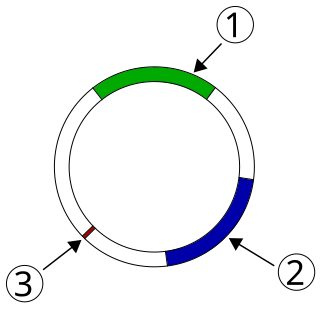
Plasmid-mediated resistance is the transfer of antibiotic resistance genes which are carried on plasmids. Plasmids possess mechanisms that ensure their independent replication as well as those that regulate their replication number and guarantee stable inheritance during cell division. By the conjugation process, they can stimulate lateral transfer between bacteria from various genera and kingdoms. Numerous plasmids contain addiction-inducing systems that are typically based on toxin-antitoxin factors and capable of killing daughter cells that don't inherit the plasmid during cell division. Plasmids often carry multiple antibiotic resistance genes, contributing to the spread of multidrug-resistance (MDR). Antibiotic resistance mediated by MDR plasmids severely limits the treatment options for the infections caused by Gram-negative bacteria, especially family Enterobacteriaceae. The global spread of MDR plasmids has been enhanced by selective pressure from antimicrobial medications used in medical facilities and when raising animals for food.
Membrane dipeptidase (EC 3.4.13.19, renal dipeptidase, dehydropeptidase I (DPH I), dipeptidase, aminodipeptidase, dipeptide hydrolase, dipeptidyl hydrolase, nonspecific dipeptidase, glycosyl-phosphatidylinositol-anchored renal dipeptidase, MBD, MDP, leukotriene D4 hydrolase) is an enzyme. This enzyme catalyses the following chemical reaction
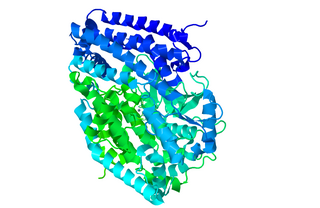
Peptidyl-dipeptidase Dcp (EC 3.4.15.5, dipeptidyl carboxypeptidase (Dcp), dipeptidyl carboxypeptidase) is a metalloenzyme found in the cytoplasm of bacterium E. Coli responsible for the C-terminal cleavage of a variety of dipeptides and unprotected larger peptide chains. The enzyme does not hydrolyze bonds in which P1' is Proline, or both P1 and P1' are Glycine. Dcp consists of 680 amino acid residues that form into a single active monomer which aids in the intracellular degradation of peptides. Dcp coordinates to divalent zinc which sits in the pocket of the active site and is composed of four subsites: S1’, S1, S2, and S3, each subsite attracts certain amino acids at a specific position on the substrate enhancing the selectivity of the enzyme. The four subsites detect and bind different amino acid types on the substrate peptide in the P1 and P2 positions. Some metallic divalent cations such as Ni+2, Cu+2, and Zn+2 inhibit the function of the enzyme around 90%, whereas other cations such as Mn+2, Ca+2, Mg+2, and Co+2 have slight catalyzing properties, and increase the function by around 20%. Basic amino acids such as Arginine bind preferably at the S1 site, the S2 site sits deeper in the enzyme therefore is restricted to bind hydrophobic amino acids with phenylalanine in the P2 position. Dcp is divided into two subdomains (I, and II), which are the two sides of the clam shell-like structure and has a deep inner cavity where a pair of histidine residues bind to the catalytic zinc ion in the active site. Peptidyl-Dipeptidase Dcp is classified like Angiotensin-I converting enzyme (ACE) which is also a carboxypeptidase involved in blood pressure regulation, but due to structural differences and peptidase activity between these two enzymes they had to be examined separately. ACE has endopeptidase activity, whereas Dcp strictly has exopeptidase activity based on its cytoplasmic location and therefore their mechanisms of action are differentiated. Another difference between these enzymes is that the activity of Peptidyl-Dipeptidase Dcp is not enhanced in the presence of chloride anions, whereas chloride enhances ACE activity.

Natalie C. J. Strynadka FRS is a professor of Biochemistry in the Department of Biochemistry and Molecular Biology at the University of British Columbia.
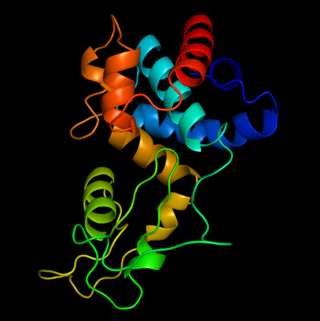
OBPgp279 is an endolysin that hydrolyzes peptidoglycan, a major constituent in bacterial membrane. OBPgp279 is found in Pseudomonas fluorescens phage OBP, which belongs in the Myoviridae family of bacteriophages. Because of its role in hydrolyzing the peptidoglycan layer, OBPgp279 is a key enzyme in the lytic cycle of the OBP bacteriophage; it allows the bacteriophage to lyse its host internally to escape. Unlike other endolysins, OBPgp279 does not rely on holins to perforate the inner bacterial membrane in order to reach the peptidoglycan layer. Although OBPgp279 is not a well-studied enzyme, it has garnered interest as a potential antibacterial protein due to its activity against multidrug-resistant gram-negative bacteria.